
An antibody (Ab) is the secreted form of a B cell receptor; the term immunoglobulin (Ig) can refer to either the membrane-bound form or the secreted form of the B cell receptor, but they are, broadly speaking, the same protein, and so the terms are often treated as synonymous. Antibodies are large, Y-shaped proteins belonging to the immunoglobulin superfamily which are used by the immune system to identify and neutralize antigens such as bacteria and viruses, including those that cause disease. Antibodies can recognize virtually any size antigen with diverse chemical compositions from molecules. Each antibody recognizes one or more specific antigens. Antigen literally means "antibody generator", as it is the presence of an antigen that drives the formation of an antigen-specific antibody. Each tip of the "Y" of an antibody contains a paratope that specifically binds to one particular epitope on an antigen, allowing the two molecules to bind together with precision. Using this mechanism, antibodies can effectively "tag" a microbe or an infected cell for attack by other parts of the immune system, or can neutralize it directly.

Immunoglobulin G (IgG) is a type of antibody. Representing approximately 75% of serum antibodies in humans, IgG is the most common type of antibody found in blood circulation. IgG molecules are created and released by plasma B cells. Each IgG antibody has two paratopes.

Immunoglobulin A is an antibody that plays a role in the immune function of mucous membranes. The amount of IgA produced in association with mucosal membranes is greater than all other types of antibody combined. In absolute terms, between three and five grams are secreted into the intestinal lumen each day. This represents up to 15% of total immunoglobulins produced throughout the body.

Immunoglobulin D (IgD) is an antibody isotype that makes up about 1% of proteins in the plasma membranes of immature B-lymphocytes where it is usually co-expressed with another cell surface antibody called IgM. IgD is also produced in a secreted form that is found in very small amounts in blood serum, representing 0.25% of immunoglobulins in serum. The relative molecular mass and half-life of secreted IgD is 185 kDa and 2.8 days, respectively. Secreted IgD is produced as a monomeric antibody with two heavy chains of the delta (δ) class, and two Ig light chains.

In molecular biology, CD4 is a glycoprotein that serves as a co-receptor for the T-cell receptor (TCR). CD4 is found on the surface of immune cells such as helper T cells, monocytes, macrophages, and dendritic cells. It was discovered in the late 1970s and was originally known as leu-3 and T4 before being named CD4 in 1984. In humans, the CD4 protein is encoded by the CD4 gene.

The T-cell receptor (TCR) is a protein complex found on the surface of T cells, or T lymphocytes, that is responsible for recognizing fragments of antigen as peptides bound to major histocompatibility complex (MHC) molecules. The binding between TCR and antigen peptides is of relatively low affinity and is degenerate: that is, many TCRs recognize the same antigen peptide and many antigen peptides are recognized by the same TCR.

The immunoglobulin heavy chain (IgH) is the large polypeptide subunit of an antibody (immunoglobulin). In human genome, the IgH gene loci are on chromosome 14.

In immunology, an Fc receptor is a protein found on the surface of certain cells – including, among others, B lymphocytes, follicular dendritic cells, natural killer cells, macrophages, neutrophils, eosinophils, basophils, human platelets, and mast cells – that contribute to the protective functions of the immune system. Its name is derived from its binding specificity for a part of an antibody known as the Fc region. Fc receptors bind to antibodies that are attached to infected cells or invading pathogens. Their activity stimulates phagocytic or cytotoxic cells to destroy microbes, or infected cells by antibody-mediated phagocytosis or antibody-dependent cell-mediated cytotoxicity. Some viruses such as flaviviruses use Fc receptors to help them infect cells, by a mechanism known as antibody-dependent enhancement of infection.

The B-cell receptor (BCR) is a transmembrane protein on the surface of a B cell. A B-cell receptor is composed of a membrane-bound immunoglobulin molecule and a signal transduction moiety. The former forms a type 1 transmembrane receptor protein, and is typically located on the outer surface of these lymphocyte cells. Through biochemical signaling and by physically acquiring antigens from the immune synapses, the BCR controls the activation of the B cell. B cells are able to gather and grab antigens by engaging biochemical modules for receptor clustering, cell spreading, generation of pulling forces, and receptor transport, which eventually culminates in endocytosis and antigen presentation. B cells' mechanical activity adheres to a pattern of negative and positive feedbacks that regulate the quantity of removed antigen by manipulating the dynamic of BCR–antigen bonds directly. Particularly, grouping and spreading increase the relation of antigen with BCR, thereby proving sensitivity and amplification. On the other hand, pulling forces delinks the antigen from the BCR, thus testing the quality of antigen binding.

Protein A is a 42 kDa surface protein originally found in the cell wall of the bacteria Staphylococcus aureus. It is encoded by the spa gene and its regulation is controlled by DNA topology, cellular osmolarity, and a two-component system called ArlS-ArlR. It has found use in biochemical research because of its ability to bind immunoglobulins. It is composed of five homologous Ig-binding domains that fold into a three-helix bundle. Each domain is able to bind proteins from many mammalian species, most notably IgGs. It binds the heavy chain within the Fc region of most immunoglobulins and also within the Fab region in the case of the human VH3 family. Through these interactions in serum, where IgG molecules are bound in the wrong orientation, the bacteria disrupts opsonization and phagocytosis.

The immunoglobulin superfamily (IgSF) is a large protein superfamily of cell surface and soluble proteins that are involved in the recognition, binding, or adhesion processes of cells. Molecules are categorized as members of this superfamily based on shared structural features with immunoglobulins ; they all possess a domain known as an immunoglobulin domain or fold. Members of the IgSF include cell surface antigen receptors, co-receptors and co-stimulatory molecules of the immune system, molecules involved in antigen presentation to lymphocytes, cell adhesion molecules, certain cytokine receptors and intracellular muscle proteins. They are commonly associated with roles in the immune system. Otherwise, the sperm-specific protein IZUMO1, a member of the immunoglobulin superfamily, has also been identified as the only sperm membrane protein essential for sperm-egg fusion.

Complementarity-determining regions (CDRs) are polypeptide segments of the variable chains in immunoglobulins (antibodies) and T cell receptors, generated by B-cells and T-cells respectively. CDRs are where these molecules bind to their specific antigen and their structure/sequence determines the binding activity of the respective antibody. A set of CDRs constitutes a paratope, or the antigen-binding site. As the most variable parts of the molecules, CDRs are crucial to the diversity of antigen specificities generated by lymphocytes.

Immunoglobulin class switching, also known as isotype switching, isotypic commutation or class-switch recombination (CSR), is a biological mechanism that changes a B cell's production of immunoglobulin from one type to another, such as from the isotype IgM to the isotype IgG. During this process, the constant-region portion of the antibody heavy chain is changed, but the variable region of the heavy chain stays the same. Since the variable region does not change, class switching does not affect antigen specificity. Instead, the antibody retains affinity for the same antigens, but can interact with different effector molecules.

In immunology, antibodies are classified into several types called isotypes or classes. The variable (V) regions near the tip of the antibody can differ from molecule to molecule in countless ways, allowing it to specifically target an antigen . In contrast, the constant (C) regions only occur in a few variants, which define the antibody's class. Antibodies of different classes activate distinct effector mechanisms in response to an antigen . They appear at different stages of an immune response, differ in structural features, and in their location around the body.

The Joining (J) chain is a protein component that links monomers of antibodies IgM and IgA to form polymeric antibodies capable of secretion. The J chain is well conserved in the animal kingdom, but its specific functions are yet to be fully understood. It is a 137 residue polypeptide, encoded by the IGJ gene.

Polymeric immunoglobulin receptor (pIgR) is a transmembrane protein that in humans is encoded by the PIGR gene. It is an Fc receptor which facilitates the transcytosis of the soluble polymeric isoforms of immunoglobulin A and immunoglobulin M (pIg) and immune complexes. pIgRs are mainly located on the epithelial lining of mucosal surfaces of the gastrointestinal tract. The composition of the receptor is complex, including 6 immunoglobulin-like domains, a transmembrane region, and an intracellular domain. pIgR expression is under the strong regulation of cytokines, hormones, and pathogenic stimuli.

Ig epsilon chain C region is a protein that in humans is encoded by the IGHE gene.

Cluster of differentiation CD79A also known as B-cell antigen receptor complex-associated protein alpha chain and MB-1 membrane glycoprotein, is a protein that in humans is encoded by the CD79A gene.

Fc fragment of IgA receptor (FCAR) is a human gene that codes for the transmembrane receptor FcαRI, also known as CD89. FcαRI binds the heavy-chain constant region of Immunoglobulin A (IgA) antibodies. FcαRI is present on the cell surface of myeloid lineage cells, including neutrophils, monocytes, macrophages, and eosinophils, though it is notably absent from intestinal macrophages and does not appear on mast cells. FcαRI plays a role in both pro- and anti-inflammatory responses depending on the state of IgA bound. Inside-out signaling primes FcαRI in order for it to bind its ligand, while outside-in signaling caused by ligand binding depends on FcαRI association with the Fc receptor gamma chain.
A heavy-chain antibody is an antibody which consists only of two heavy chains and lacks the two light chains usually found in antibodies.
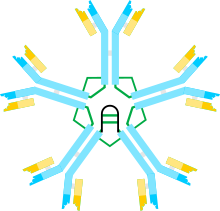

![Figure 2. Some alternative ways of linking m chains
A, B) These figures depict two of many possible models of inter-m chain disulfide bonding in hexameric IgM. As in Figure 1, the Cm2 disulfide bonds and the Cm4tp disulfide bonds are represented by a red double arrowhead, and the Cm3 disulfide bonds are represented by the long double-headed arrows. In both models A and B each type of disulfide bond (Cm2-Cm2; Cm3-Cm3; Cm4tp-Cm4tp) joins m chains eries with each of the others. Methods for distinguishing these and other models are discussed in reference [28].
C) This representation of pentameric IgM illustrates how the J chain might be bonded to m chains that are not linked via Cm3 disulfide bonds Some alternative ways of linking u chains.jpg](http://upload.wikimedia.org/wikipedia/commons/thumb/0/02/Some_alternative_ways_of_linking_%C2%B5_chains.jpg/220px-Some_alternative_ways_of_linking_%C2%B5_chains.jpg)

















