Related Research Articles

In biology, epigenetics is the study of heritable traits, or a stable change of cell function, that happen without changes to the DNA sequence. The Greek prefix epi- in epigenetics implies features that are "on top of" or "in addition to" the traditional genetic mechanism of inheritance. Epigenetics usually involves a change that is not erased by cell division, and affects the regulation of gene expression. Such effects on cellular and physiological phenotypic traits may result from environmental factors, or be part of normal development. They can lead to cancer.
Methylation, in the chemical sciences, is the addition of a methyl group on a substrate, or the substitution of an atom by a methyl group. Methylation is a form of alkylation, with a methyl group replacing a hydrogen atom. These terms are commonly used in chemistry, biochemistry, soil science, and biology.

5-Methylcytosine is a methylated form of the DNA base cytosine (C) that regulates gene transcription and takes several other biological roles. When cytosine is methylated, the DNA maintains the same sequence, but the expression of methylated genes can be altered. 5-Methylcytosine is incorporated in the nucleoside 5-methylcytidine.

The CpG sites or CG sites are regions of DNA where a cytosine nucleotide is followed by a guanine nucleotide in the linear sequence of bases along its 5' → 3' direction. CpG sites occur with high frequency in genomic regions called CpG islands.

In biochemistry, the DNA methyltransferase family of enzymes catalyze the transfer of a methyl group to DNA. DNA methylation serves a wide variety of biological functions. All the known DNA methyltransferases use S-adenosyl methionine (SAM) as the methyl donor.

Histone methyltransferases (HMT) are histone-modifying enzymes, that catalyze the transfer of one, two, or three methyl groups to lysine and arginine residues of histone proteins. The attachment of methyl groups occurs predominantly at specific lysine or arginine residues on histones H3 and H4. Two major types of histone methyltranferases exist, lysine-specific and arginine-specific. In both types of histone methyltransferases, S-Adenosyl methionine (SAM) serves as a cofactor and methyl donor group.
The genomic DNA of eukaryotes associates with histones to form chromatin. The level of chromatin compaction depends heavily on histone methylation and other post-translational modifications of histones. Histone methylation is a principal epigenetic modification of chromatin that determines gene expression, genomic stability, stem cell maturation, cell lineage development, genetic imprinting, DNA methylation, and cell mitosis.

DNA methylation is a biological process by which methyl groups are added to the DNA molecule. Methylation can change the activity of a DNA segment without changing the sequence. When located in a gene promoter, DNA methylation typically acts to repress gene transcription. In mammals, DNA methylation is essential for normal development and is associated with a number of key processes including genomic imprinting, X-chromosome inactivation, repression of transposable elements, aging, and carcinogenesis.

In organic chemistry, a methoxy group is the functional group consisting of a methyl group bound to oxygen. This alkoxy group has the formula R−O−CH3.
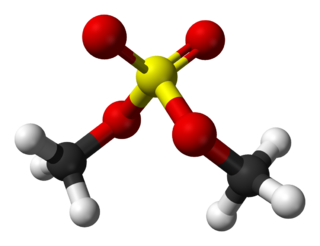
Dimethyl sulfate (DMS) is a chemical compound with formula (CH3O)2SO2. As the diester of methanol and sulfuric acid, its formula is often written as (CH3)2SO4 or Me2SO4, where CH3 or Me is methyl. Me2SO4 is mainly used as a methylating agent in organic synthesis.
Histone methylation is a process by which methyl groups are transferred to amino acids of histone proteins that make up nucleosomes, which the DNA double helix wraps around to form chromosomes. Methylation of histones can either increase or decrease transcription of genes, depending on which amino acids in the histones are methylated, and how many methyl groups are attached. Methylation events that weaken chemical attractions between histone tails and DNA increase transcription because they enable the DNA to uncoil from nucleosomes so that transcription factor proteins and RNA polymerase can access the DNA. This process is critical for the regulation of gene expression that allows different cells to express different genes.

Methyltransferases are a large group of enzymes that all methylate their substrates but can be split into several subclasses based on their structural features. The most common class of methyltransferases is class I, all of which contain a Rossmann fold for binding S-Adenosyl methionine (SAM). Class II methyltransferases contain a SET domain, which are exemplified by SET domain histone methyltransferases, and class III methyltransferases, which are membrane associated. Methyltransferases can also be grouped as different types utilizing different substrates in methyl transfer reactions. These types include protein methyltransferases, DNA/RNA methyltransferases, natural product methyltransferases, and non-SAM dependent methyltransferases. SAM is the classical methyl donor for methyltransferases, however, examples of other methyl donors are seen in nature. The general mechanism for methyl transfer is a SN2-like nucleophilic attack where the methionine sulfur serves as the leaving group and the methyl group attached to it acts as the electrophile that transfers the methyl group to the enzyme substrate. SAM is converted to S-Adenosyl homocysteine (SAH) during this process. The breaking of the SAM-methyl bond and the formation of the substrate-methyl bond happen nearly simultaneously. These enzymatic reactions are found in many pathways and are implicated in genetic diseases, cancer, and metabolic diseases. Another type of methyl transfer is the radical S-Adenosyl methionine (SAM) which is the methylation of unactivated carbon atoms in primary metabolites, proteins, lipids, and RNA.

A catechol-O-methyltransferase(COMT) inhibitor is a drug that inhibits the enzyme catechol-O-methyltransferase. This enzyme methylates catecholamines such as dopamine, norepinephrine and epinephrine. It also methylates levodopa. COMT inhibitors are indicated for the treatment of Parkinson's disease in combination with levodopa and an aromatic L-amino acid decarboxylase inhibitor. The therapeutic benefit of using a COMT inhibitor is based on its ability to prevent the methylation of levodopa to 3-O-methyldopa, thus increasing the bioavailability of levodopa. COMT inhibitors significantly decrease off time in people with Parkinson's disease also taking carbidopa/levodopa.

For molecular biology in mammals, DNA demethylation causes replacement of 5-methylcytosine (5mC) in a DNA sequence by cytosine (C). DNA demethylation can occur by an active process at the site of a 5mC in a DNA sequence or, in replicating cells, by preventing addition of methyl groups to DNA so that the replicated DNA will largely have cytosine in the DNA sequence.

Petunidin (Pt), like Europinidin and Malvidin, is derived from Delphinidin and is an O-methylated anthocyanidin of the 3-hydroxy type. It is a natural organic compound, a dark-red or purple water-soluble pigment found in many red berries including chokeberries, Saskatoon berries or different species of grape, and also part of the pigments responsible for the petal colors in many flowers. This pigment gives the Indigo Rose tomatoes the majority of their deep purple color when the fruits are exposed to sunlight. The name of the molecule itself is derived from the word Petunia.

Tectorigenin is an O-methylated isoflavone, a type of flavonoid. It ha been isolated from leopard lily, Algerian iris and East Asian arrowroot.
The O-methylated flavonoids or methoxyflavonoids are flavonoids with methylations on hydroxyl groups. O-methylation has an effect on the solubility of flavonoids.

Ayanin is an O-methylated flavonol, a type of flavonoid. It is the 3,7,4'-tri-O-methylated derivative of quercetin.

Ombuin is an O-methylated flavonol, a type of flavonoid. It is the 4',7-O-methyl derivative of quercetin.
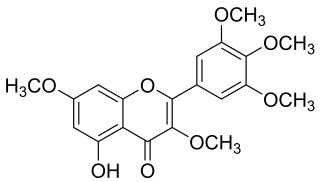
Combretol is an O-methylated flavonol, a type of flavonoid. It is the 3,7,3',4',5'-O-methylation of myricetin and can be extracted from Combretum quadrangulare and from Rhodomyrtus tomentosa.

Bayesian tool for methylation analysis, also known as BATMAN, is a statistical tool for analysing methylated DNA immunoprecipitation (MeDIP) profiles. It can be applied to large datasets generated using either oligonucleotide arrays (MeDIP-chip) or next-generation sequencing (MeDIP-seq), providing a quantitative estimation of absolute methylation state in a region of interest.
References
- ↑ Koirala, Niranjan; Thuan, Nguyen Huy; Ghimire, Gopal Prasad; Thang, Duong Van; Sohng, Jae Kyung (2016-02-23). "Methylation of flavonoids: Chemical structures, bioactivities, progress and perspectives for biotechnological production". Enzyme and Microbial Technology . 86: 103–116. doi:10.1016/j.enzmictec.2016.02.003 – via ScienceDirect.