
Cytochromes are redox-active proteins containing a heme, with a central Fe atom at its core, as a cofactor. They are involved in electron transport chain and redox catalysis. They are classified according to the type of heme and its mode of binding. Four varieties are recognized by the International Union of Biochemistry and Molecular Biology (IUBMB), cytochromes a, cytochromes b, cytochromes c and cytochrome d. Cytochrome function is linked to the reversible redox change from ferrous to the ferric oxidation state of the iron found in the heme core. In addition to the classification by the IUBMB into four cytochrome classes, several additional classifications such as cytochrome o and cytochrome P450 can be found in biochemical literature.

Oxidative phosphorylation or electron transport-linked phosphorylation or terminal oxidation is the metabolic pathway in which cells use enzymes to oxidize nutrients, thereby releasing the chemical energy stored within the nutrients in order to produce adenosine triphosphate (ATP). In eukaryotes, this takes place inside mitochondria. Almost all aerobic organisms carry out oxidative phosphorylation. This pathway is so pervasive because it releases more energy than alternative fermentation processes such as anaerobic glycolysis.

The electron transport chain (ETC; respiratory chain) is a series of protein complexes that transfer electrons from electron donors to electron acceptors via redox reactions (both reduction and oxidation occurring simultaneously) and couples this electron transfer with the transfer of protons (H+ ions) across a membrane. The electron transport chain is built up of peptides, enzymes, and other molecules.

The enzyme cytochrome c oxidase or Complex IV, EC 1.9.3.1, is a large transmembrane protein complex found in bacteria, archaea, and the mitochondria of eukaryotes.
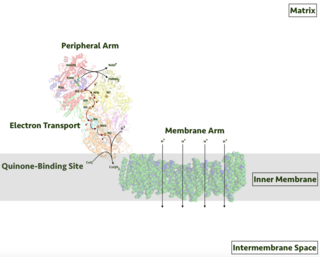
Respiratory complex I, EC 7.1.1.2 is the first large protein complex of the respiratory chains of many organisms from bacteria to humans. It catalyzes the transfer of electrons from NADH to coenzyme Q10 (CoQ10) and translocates protons across the inner mitochondrial membrane in eukaryotes or the plasma membrane of bacteria.
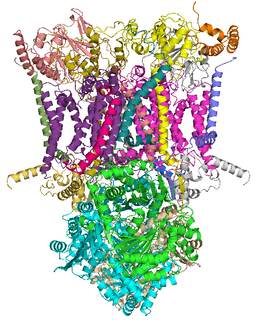
The coenzyme Q : cytochrome c – oxidoreductase, sometimes called the cytochrome bc1 complex, and at other times complex III, is the third complex in the electron transport chain, playing a critical role in biochemical generation of ATP. Complex III is a multisubunit transmembrane protein encoded by both the mitochondrial and the nuclear genomes. Complex III is present in the mitochondria of all animals and all aerobic eukaryotes and the inner membranes of most eubacteria. Mutations in Complex III cause exercise intolerance as well as multisystem disorders. The bc1 complex contains 11 subunits, 3 respiratory subunits, 2 core proteins and 6 low-molecular weight proteins.

Succinate dehydrogenase (SDH) or succinate-coenzyme Q reductase (SQR) or respiratory complex II is an enzyme complex, found in many bacterial cells and in the inner mitochondrial membrane of eukaryotes. It is the only enzyme that participates in both the citric acid cycle and the electron transport chain. Histochemical analysis showing high succinate dehydrogenase in muscle demonstrates high mitochondrial content and high oxidative potential.

Rieske proteins are iron–sulfur protein (ISP) components of cytochrome bc1 complexes and cytochrome b6f complexes and are responsible for electron transfer in some biological systems. John S. Rieske and co-workers first discovered the protein and in 1964 isolated an acetylated form of the bovine mitochondrial protein. In 1979 Trumpower's lab isolated the "oxidation factor" from bovine mitochondria and showed it was a reconstitutively-active form of the Rieske iron-sulfur protein
It is a unique [2Fe-2S] cluster in that one of the two Fe atoms is coordinated by two histidine residues rather than two cysteine residues. They have since been found in plants, animals, and bacteria with widely ranging electron reduction potentials from -150 to +400 mV.

The Q cycle describes a series of reactions that describe how the sequential oxidation and reduction of the lipophilic electron carrier, Coenzyme Q10 (CoQ10), between the ubiquinol and ubiquinone forms, can result in the net movement of protons across a lipid bilayer.
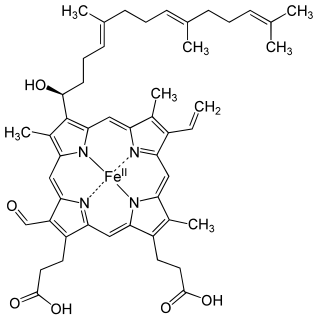
Heme A is a heme, a coordination complex consisting of a macrocyclic ligand called a porphyrin, chelating an iron atom. Heme A is a biomolecule and is produced naturally by many organisms. Heme A, often appears a dichroic green/red when in solution, is a structural relative of heme B, a component of hemoglobin, the red pigment in blood.
Ubiquinol oxidases are enzymes in the bacterial electron transport chain that oxidise ubiquinol into ubiquinone and reduce oxygen to water. These enzymes are one set of the many alternative terminal oxidases in the branched prokaryotic electron transport chain. The overall structure of the E. coli ubiquinol oxidase is similar to that of the mammalian Cytochrome c oxidase, with the addition of a polar ubiquinol-binding site embedded in the membrane.

Cytochrome c oxidase I (COX1) also known as mitochondrially encoded cytochrome c oxidase I (MT-CO1) is a protein that in humans is encoded by the MT-CO1 gene. In other eukaryotes, the gene is called COX1, CO1, or COI. Cytochrome c oxidase I is the main subunit of the cytochrome c oxidase complex. Mutations in MT-CO1 have been associated with Leber's hereditary optic neuropathy (LHON), acquired idiopathic sideroblastic anemia, Complex IV deficiency, colorectal cancer, sensorineural deafness, and recurrent myoglobinuria.

Cytochrome c oxidase subunit III (COX3) is an enzyme that in humans is encoded by the MT-CO3 gene. It is one of main transmembrane subunits of cytochrome c oxidase. Cytochrome c oxidase subunit III is also one of the three mitochondrial DNA (mtDNA) encoded subunits of respiratory complex IV. Variants of MT-CO3 have been associated with isolated myopathy, severe encephalomyopathy, Leber hereditary optic neuropathy, mitochondrial complex IV deficiency, and recurrent myoglobinuria.

Cytochrome c oxidase subunit 5B, mitochondrial is an enzyme in humans that is a subunit of the cytochrome c oxidase complex, also known as Complex IV, the last enzyme in the mitochondrial electron transport chain. In humans, cytochrome c oxidase subunit 5B is encoded by the COX5B gene.

Ubiquinol-cytochrome c reductase binding protein, also known as UQCRB, Complex III subunit 7, QP-C, or Ubiquinol-cytochrome c reductase complex 14 kDa protein is a protein which in humans is encoded by the UQCRB gene. This gene encodes a subunit of the ubiquinol-cytochrome c oxidoreductase complex, which consists of one mitochondrial-encoded and 10 nuclear-encoded subunits. Mutations in this gene are associated with mitochondrial complex III deficiency. Alternatively spliced transcript variants have been found for this gene. Related pseudogenes have been identified on chromosomes 1, 5 and X.

Fumarate reductase (quinol) is an enzyme with systematic name succinate:quinone oxidoreductase. This enzyme catalyzes the following chemical reaction:
Pyruvate dehydrogenase (quinone) (EC 1.2.5.1, pyruvate dehydrogenase, pyruvic dehydrogenase, pyruvic (cytochrome b1) dehydrogenase, pyruvate:ubiquinone-8-oxidoreductase, pyruvate oxidase (ambiguous)) is an enzyme with systematic name pyruvate:ubiquinone oxidoreductase. This enzyme catalyses the following chemical reaction
Ubiquinol oxidase (H+-transporting) (EC 7.1.1.3, cytochrome bb3 oxidase, cytochrome bo oxidase, cytochrome bd-I oxidase) is an enzyme with systematic name ubiquinol:O2 oxidoreductase (H+-transporting). This enzyme catalyses the following chemical reaction

Cytochrome c oxidase subunit 8A (COX8A) is a protein that in humans is encoded by the COX8A gene. Cytochrome c oxidase 8A is a subunit of the cytochrome c oxidase complex, also known as Complex IV. Mutations in the COX8A gene have been associated with complex IV deficiency with Leigh syndrome and epilepsy.

NDUFA4, mitochondrial complex associated is a protein that in humans is encoded by the NDUFA4 gene. The NDUFA3 protein is a subunit of NADH dehydrogenase (ubiquinone), which is located in the mitochondrial inner membrane and is the largest of the five complexes of the electron transport chain. Mutations in the NDUFA4 gene are associated with Leigh's syndrome.
















