
Enzymes are proteins that act as biological catalysts by accelerating chemical reactions. The molecules upon which enzymes may act are called substrates, and the enzyme converts the substrates into different molecules known as products. Almost all metabolic processes in the cell need enzyme catalysis in order to occur at rates fast enough to sustain life. Metabolic pathways depend upon enzymes to catalyze individual steps. The study of enzymes is called enzymology and the field of pseudoenzyme analysis recognizes that during evolution, some enzymes have lost the ability to carry out biological catalysis, which is often reflected in their amino acid sequences and unusual 'pseudocatalytic' properties.

Oxidative phosphorylation or electron transport-linked phosphorylation or terminal oxidation is the metabolic pathway in which cells use enzymes to oxidize nutrients, thereby releasing chemical energy in order to produce adenosine triphosphate (ATP). In eukaryotes, this takes place inside mitochondria. Almost all aerobic organisms carry out oxidative phosphorylation. This pathway is so pervasive because it releases more energy than alternative fermentation processes such as anaerobic glycolysis.

An electron transport chain (ETC) is a series of protein complexes and other molecules that transfer electrons from electron donors to electron acceptors via redox reactions (both reduction and oxidation occurring simultaneously) and couples this electron transfer with the transfer of protons (H+ ions) across a membrane. The electrons that transferred from NADH and FADH2 to the ETC involves 4 multi-subunit large enzymes complexes and 2 mobile electron carriers. Many of the enzymes in the electron transport chain are membrane-bound.

The enzyme cytochrome c oxidase or Complex IV, is a large transmembrane protein complex found in bacteria, archaea, and mitochondria of eukaryotes.

Heme, or haem, is a precursor to hemoglobin, which is necessary to bind oxygen in the bloodstream. Heme is biosynthesized in both the bone marrow and the liver.
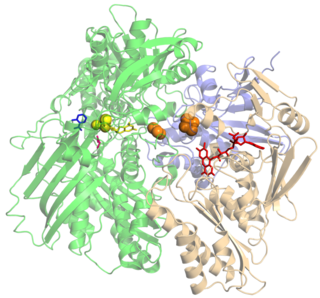
Xanthine oxidase is a form of xanthine oxidoreductase, a type of enzyme that generates reactive oxygen species. These enzymes catalyze the oxidation of hypoxanthine to xanthine and can further catalyze the oxidation of xanthine to uric acid. These enzymes play an important role in the catabolism of purines in some species, including humans.

Cytochromes P450 (CYPs) are a superfamily of enzymes containing heme as a cofactor that function as monooxygenases. In mammals, these proteins oxidize steroids, fatty acids, and xenobiotics, and are important for the clearance of various compounds, as well as for hormone synthesis and breakdown. In 1963, Estabrook, Cooper, and Rosenthal described the role of CYP as a catalyst in steroid hormone synthesis and drug metabolism. In plants, these proteins are important for the biosynthesis of defensive compounds, fatty acids, and hormones.
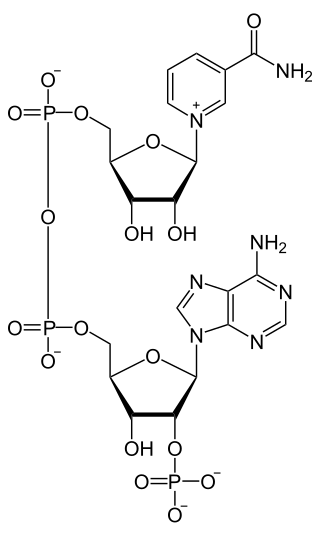
Nicotinamide adenine dinucleotide phosphate, abbreviated NADP+ or, in older notation, TPN (triphosphopyridine nucleotide), is a cofactor used in anabolic reactions, such as the Calvin cycle and lipid and nucleic acid syntheses, which require NADPH as a reducing agent ('hydrogen source'). It is used by all forms of cellular life.
Drug metabolism is the metabolic breakdown of drugs by living organisms, usually through specialized enzymatic systems. More generally, xenobiotic metabolism is the set of metabolic pathways that modify the chemical structure of xenobiotics, which are compounds foreign to an organism's normal biochemistry, such as any drug or poison. These pathways are a form of biotransformation present in all major groups of organisms and are considered to be of ancient origin. These reactions often act to detoxify poisonous compounds. The study of drug metabolism is called pharmacokinetics.
NADPH oxidase is a membrane-bound enzyme complex that faces the extracellular space. It can be found in the plasma membrane as well as in the membranes of phagosomes used by neutrophil white blood cells to engulf microorganisms. Human isoforms of the catalytic component of the complex include NOX1, NOX2, NOX3, NOX4, NOX5, DUOX1, and DUOX2.
Any enzyme system that includes cytochrome P450 protein or domain can be called a P450-containing system.
Microbial metabolism is the means by which a microbe obtains the energy and nutrients it needs to live and reproduce. Microbes use many different types of metabolic strategies and species can often be differentiated from each other based on metabolic characteristics. The specific metabolic properties of a microbe are the major factors in determining that microbe's ecological niche, and often allow for that microbe to be useful in industrial processes or responsible for biogeochemical cycles.

Acetohydroxamic acid is a drug that is a potent and irreversible enzyme inhibitor of the urease enzyme in various bacteria and plants; it is usually used for urinary tract infections. The molecule is similar to urea but is not hydrolyzable by urease; it thus disrupts the bacteria's metabolism through competitive inhibition.

Aldehyde oxidase (AO) is a metabolizing enzyme, located in the cytosolic compartment of tissues in many organisms. AO catalyzes the oxidation of aldehydes into carboxylic acid, and in addition, catalyzes the hydroxylation of some heterocycles. It can also catalyze the oxidation of both cytochrome P450 (CYP450) and monoamine oxidase (MAO) intermediate products. AO plays an important role in the metabolism of several drugs.

Squalene monooxygenase is a eukaryotic enzyme that uses NADPH and diatomic oxygen to oxidize squalene to 2,3-oxidosqualene. Squalene epoxidase catalyzes the first oxygenation step in sterol biosynthesis and is thought to be one of the rate-limiting enzymes in this pathway. In humans, squalene epoxidase is encoded by the SQLE gene. Several eukaryote genomes lack a squalene monooxygenase encoding gene, but instead encode an alternative squalene epoxidase that performs the same task.
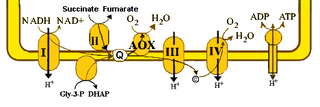
The alternative oxidase (AOX) is an enzyme that forms part of the electron transport chain in mitochondria of different organisms. Proteins homologous to the mitochondrial oxidase and the related plastid terminal oxidase have also been identified in bacterial genomes.

Ascofuranone is an antibiotic produced by various ascomycete fungi including Acremonium sclerotigenum that inhibits the Trypanosoma brucei alternative oxidase and is a lead compound in efforts to produce other drugs targeting this enzyme for the treatment of sleeping sickness. The compound is effective both in vitro cell culture and in infections in mice.

Diphosphomevalonate decarboxylase (EC 4.1.1.33), most commonly referred to in scientific literature as mevalonate diphosphate decarboxylase, is an enzyme that catalyzes the chemical reaction

Azurin is a small, periplasmic, bacterial blue copper protein found in Pseudomonas, Bordetella, or Alcaligenes bacteria. Azurin moderates single-electron transfer between enzymes associated with the cytochrome chain by undergoing oxidation-reduction between Cu(I) and Cu(II). Each monomer of an azurin tetramer has a molecular weight of approximately 14kDa, contains a single copper atom, is intensively blue, and has a fluorescence emission band centered at 308 nm.
The Arc system is a two-component system found in some bacteria that regulates gene expression in faculatative anaerobes such as Escheria coli. Two-component system means that it has a sensor molecule and a response regulator. Arc is an abbreviation for Anoxic Redox Control system. Arc systems are instrumental in maintaining energy metabolism during transcription of bacteria. The ArcA response regulator looks at growth conditions and expresses genes to best suit the bacteria. The Arc B sensor kinase, which is a tripartite protein, is membrane bound and can autophosphorylate.














