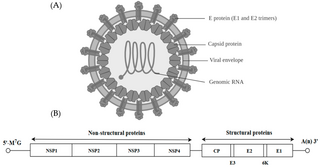
A viral envelope is the outermost layer of many types of viruses. [1] It protects the genetic material in their life cycle when traveling between host cells. Not all viruses have envelopes. A viral envelope protein or E protein is a protein in the envelope, which may be acquired by the capsid from an infected host cell.
Contents
Numerous human pathogenic viruses in circulation are encased in lipid bilayers, and they infect their target cells by causing the viral envelope and cell membrane to fuse. Although there are effective vaccines against some of these viruses, there is no preventative or curative medicine for the majority of them. In most cases, the known vaccines operate by inducing antibodies that prevent the pathogen from entering cells. This happens in the case of enveloped viruses when the antibodies bind to the viral envelope proteins.
The membrane fusion event that triggers viral entrance is caused by the viral fusion protein. Many enveloped viruses only have one protein visible on the surface of the particle, which is required for both mediating adhesion to the cell surface and for the subsequent membrane fusion process. To create potentially protective vaccines for human pathogenic enveloped viruses for which there is currently no vaccine, it is essential to comprehend how antibodies interact with viral envelope proteins, particularly with the fusion protein, and how antibodies neutralize viruses.
Enveloped viruses enter cells by joining a cellular membrane to their lipid bilayer membrane. Priming by proteolytic processing, either of the fusion protein or of a companion protein, is necessary for the majority of viral fusion proteins. The priming stage then gets the fusion protein ready for triggering by the processes that go along with attachment and uptake, which frequently happens during transport of the fusion protein to the cell surface but may also happen extracellularly. So far, structural studies have revealed two kinds of viral fusion proteins. These proteins are believed to catalyze the same mechanism in both situations, resulting in the fusing of two bilayers. In other words, these proteins operate as enzymes, which while having various structural variations catalyze the same chemical reaction. [2]
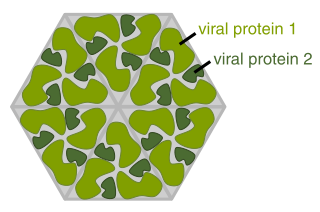
The envelopes are typically derived from portions of the host cell membranes (phospholipids and proteins), but include some viral glycoproteins. One of the main parts of human pathogenic viruses is glycoprotein. They have been shown to play significant roles in immunity and infection. [3] Viral glycoproteins, a new class of cellular inhibitory proteins has been discovered. These include the E3 ubiquitin ligases of the membrane-associated RING-CH (MARCH) family, which among other things, inhibits the expression of cell surface proteins implicated in adaptive immunity. [4] Being made up mostly of host membrane, the viral envelope can also have the proteins associated with the host cell within their membrane after budding. [5] Many enveloped viruses mature by budding at the plasma membrane, which allows them to be discharged from infected cells. During this procedure, viral transmembrane proteins, also known as spike proteins, are integrated into membrane vesicles containing components of the viral core (capsid).
For a very long time, it was thought that the spike proteins, which are necessary for infectivity, were directly incorporated into the viral core through their cytoplasmic domains. Recent research suggests that while such direct interactions may be what causes the budding of alphaviruses, this may not be the case for retroviruses and negative strand RNA viruses. These viruses can form bud particles even in the absence of spike proteins by relying only on viral core components. The spike proteins can occasionally be produced as virus-like particles without the viral core. Therefore, optimal budding and release may be dependent on a coordinated "push-and-pull" action between core and spike, where oligomerization of both components is essential. [6]
They may help viruses avoid the host immune system. TAM receptor tyrosine kinases increase phagocytic clearance of apoptotic cells and inhibit immunological responses brought on by Toll-like receptors and type I interferons (IFNs) when they are activated by the ligands Gas6 and Protein S. The phospholipid phosphatidylserine may be seen on the membranes of several enveloped viruses, which they employ to bind Gas6 and Protein S to activate TAM receptors.
Ligand-coated viruses stimulate type I IFN signaling, activate TAM receptors on dendritic cells (DCs), and suppress type II interferon signaling to circumvent host defenses and advance infection.TAM-deficient DCs exhibit type I IFN responses that are more pronounced than those of wild-type cells in response to viral exposure. As a result, flaviviruses and pseudo typed retroviruses have a harder time infecting TAM-deficient DCs, albeit infection can be brought back by type I IFN antibodies. A TAM kinase inhibitor, meanwhile, prevents infection of wild-type DCs. TAM receptors, which are potential targets for therapy, are thereby activated by viruses to reduce type I IFN signaling. [7] Glycoproteins on the surface of the envelope serve to identify and bind to receptor sites on the host's membrane. The particular set of viral proteins are engaged in a series of structural changes. When these changes are set/finished, there is then and only then, fusion with the host membrane. [8] These glycoproteins mediate the interaction between virion and host cell, typically initiating the fusion between the viral envelope and the host's cellular membrane. [9] In some cases, the virus with an envelope will form an endosome within the host cell. [10] There are three main types of viral glycoproteins: Envelope proteins, membrane proteins, and spike proteins (E, M, and S). [11] The viral envelope then fuses with the host's membrane, allowing the capsid and viral genome to enter and infect the host.[ citation needed ]
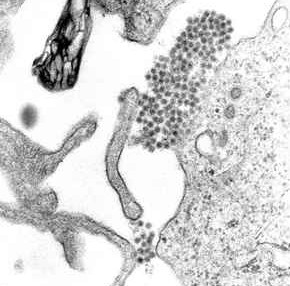
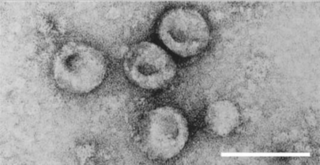
All enveloped viruses also have a capsid, another protein layer, between the envelope and the genome. [1] The virus wraps its delicate nucleic acid with a protein shell known as the capsid, from the Latin capsa, meaning "box," in order to shield it from this hostile environment. Similar to how numerous bricks come together to form a wall, the capsid is made up of one or more distinct protein types that repeatedly repeat to form the whole capsid. This repetitive pattern creates a robust but rather flexible capsid. The nucleic acid inside the capsid is appropriately protected by its modest size and physical difficulty in opening it. The nucleocapsid of the virion is made up of the nucleic acid and the capsid. Remember that the genomes of most viruses are very small. Genes code for instructions to make proteins, so small genomes cannot code for many proteins. Therefore, the virion capsid consists of one or only a few proteins that repeat over and over to form the structure. The viral nucleic acid would be physically too large to fit inside the capsid if it consisted of more than a few proteins. [12] The capsid, having a focused role of protecting the genome in addition to immune recognition evasion. [13] The viral capsid is known for its protection of RNA before it is inserted into the host cell, unlike the viral envelope which protects the protein capsid. [14]
The cell from which a virus buds often dies or is weakened, and sheds more viral particles for an extended period. The lipid bilayer envelope of these viruses is relatively sensitive to desiccation, heat, and amphiphiles such as soap and detergents, therefore these viruses are easier to sterilize than non-enveloped viruses, have limited survival outside host environments, and typically must transfer directly from host to host. Viral envelope persistence, whether it be enveloped or naked, are a factor in determining longevity of a virus on inanimate surfaces. [15] Enveloped viruses possess great adaptability and can change in a short time in order to evade the immune system. Enveloped viruses can cause persistent infections.[ citation needed ]
Vaccination against enveloped viruses can function by neutralizing the glycoprotein activity with antibodies. [16]