Coronaviruses are a group of related RNA viruses that cause diseases in mammals and birds. In humans and birds, they cause respiratory tract infections that can range from mild to lethal. Mild illnesses in humans include some cases of the common cold, while more lethal varieties can cause SARS, MERS and COVID-19. In cows and pigs they cause diarrhea, while in mice they cause hepatitis and encephalomyelitis.

Severe-acute-respiratory-syndrome–related coronavirus is a species of virus consisting of many known strains. Two strains of the virus have caused outbreaks of severe respiratory diseases in humans: severe acute respiratory syndrome coronavirus 1, which caused the 2002–2004 outbreak of severe acute respiratory syndrome (SARS), and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which is causing the ongoing pandemic of COVID-19. There are hundreds of other strains of SARSr-CoV, which are only known to infect non-human mammal species: bats are a major reservoir of many strains of SARSr-CoV; several strains have been identified in Himalayan palm civets, which were likely ancestors of SARS-CoV-1.

Coronaviridae is a family of enveloped, positive-strand RNA viruses which infect amphibians, birds, and mammals. The group includes the subfamilies Letovirinae and Orthocoronavirinae; the members of the latter are known as coronaviruses.

Human coronavirus NL63 (HCoV-NL63) is a species of coronavirus, specifically a Setracovirus from among the Alphacoronavirus genus. It was identified in late 2004 in patients in the Netherlands by Lia van der Hoek and Krzysztof Pyrc using a novel virus discovery method VIDISCA. Later on the discovery was confirmed by the researchers from the Rotterdam, the Netherlands The virus is an enveloped, positive-sense, single-stranded RNA virus which enters its host cell by binding to ACE2. Infection with the virus has been confirmed worldwide, and has an association with many common symptoms and diseases. Associated diseases include mild to moderate upper respiratory tract infections, severe lower respiratory tract infection, croup and bronchiolitis.

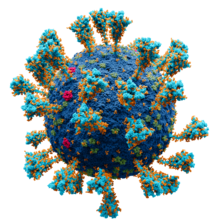
In virology, a spike protein or peplomer protein is a protein that forms a large structure known as a spike or peplomer projecting from the surface of an enveloped virus. The proteins are usually glycoproteins that form dimers or trimers.
A viral structural protein is a viral protein that is a structural component of the mature virus.
Feline coronavirus (FCoV) is a positive-stranded RNA virus that infects cats worldwide. It is a coronavirus of the species Alphacoronavirus 1, which includes canine coronavirus (CCoV) and porcine transmissible gastroenteritis coronavirus (TGEV). FCoV has two different forms: feline enteric coronavirus (FECV), which infects the intestines, and feline infectious peritonitis virus (FIPV), which causes the disease feline infectious peritonitis (FIP).

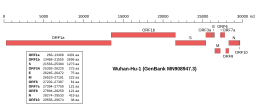
ORF7a is a gene found in coronaviruses of the Betacoronavirus genus. It expresses the Betacoronavirus NS7A protein, a type I transmembrane protein with an immunoglobulin-like protein domain. It was first discovered in SARS-CoV, the virus that causes severe acute respiratory syndrome (SARS). The homolog in SARS-CoV-2, the virus that causes COVID-19, has about 85% sequence identity to the SARS-CoV protein.

Human coronavirus HKU1 (HCoV-HKU1) is a species of coronavirus in humans and animals. It causes an upper respiratory disease with symptoms of the common cold, but can advance to pneumonia and bronchiolitis. It was first discovered in January 2004 from one man in Hong Kong. Subsequent research revealed it has global distribution and earlier genesis.

Betacoronavirus is one of four genera of coronaviruses. Member viruses are enveloped, positive-strand RNA viruses that infect mammals, including humans. The natural reservoir for betacoronaviruses are bats and rodents. Rodents are the reservoir for the subgenus Embecovirus, while bats are the reservoir for the other subgenera.

Human coronavirus 229E (HCoV-229E) is a species of coronavirus which infects humans and bats. It is an enveloped, positive-sense, single-stranded RNA virus which enters its host cell by binding to the APN receptor. Along with Human coronavirus OC43, it is one of the viruses responsible for the common cold. HCoV-229E is a member of the genus Alphacoronavirus and subgenus Duvinacovirus.
Bat SARS-like coronavirus WIV1, also sometimes called SARS-like coronavirus WIV1, is a strain of severe acute respiratory syndrome–related coronavirus (SARSr-CoV) isolated from Chinese rufous horseshoe bats in 2013. Like all coronaviruses, virions consist of single-stranded positive-sense RNA enclosed within an envelope.

The envelope (E) protein is the smallest and least well-characterized of the four major structural proteins found in coronavirus virions. It is an integral membrane protein less than 110 amino acid residues long; in SARS-CoV-2, the causative agent of Covid-19, the E protein is 75 residues long. Although it is not necessarily essential for viral replication, absence of the E protein may produce abnormally assembled viral capsids or reduced replication. E is a multifunctional protein and, in addition to its role as a structural protein in the viral capsid, it is thought to be involved in viral assembly, likely functions as a viroporin, and is involved in viral pathogenesis.

The nucleocapsid (N) protein is a protein that packages the positive-sense RNA genome of coronaviruses to form ribonucleoprotein structures enclosed within the viral capsid. The N protein is the most highly expressed of the four major coronavirus structural proteins. In addition to its interactions with RNA, N forms protein-protein interactions with the coronavirus membrane protein (M) during the process of viral assembly. N also has additional functions in manipulating the cell cycle of the host cell. The N protein is highly immunogenic and antibodies to N are found in patients recovered from SARS and COVID-19.

Spike (S) glycoprotein is the largest of the four major structural proteins found in coronaviruses. The spike protein assembles into trimers that form large structures, called spikes or peplomers, that project from the surface of the virion. The distinctive appearance of these spikes when visualized using negative stain transmission electron microscopy, "recalling the solar corona", gives the virus family its main name.

ORF3a is a gene found in coronaviruses of the subgenus Sarbecovirus, including SARS-CoV and SARS-CoV-2. It encodes an accessory protein about 275 amino acid residues long, which is thought to function as a viroporin. It is the largest accessory protein and was the first of the SARS-CoV accessory proteins to be described.
ORF7b is a gene found in coronaviruses of the genus Betacoronavirus, which expresses the accessory protein Betacoronavirus NS7b protein. It is a short, highly hydrophobic transmembrane protein of unknown function.

ORF8 is a gene that encodes a viral accessory protein, Betacoronavirus NS8 protein, in coronaviruses of the subgenus Sarbecovirus. It is one of the least well conserved and most variable parts of the genome. In some viruses, a deletion splits the region into two smaller open reading frames, called ORF8a and ORF8b - a feature present in many SARS-CoV viral isolates from later in the SARS epidemic, as well as in some bat coronaviruses. For this reason the full-length gene and its protein are sometimes called ORF8ab. The full-length gene, exemplified in SARS-CoV-2, encodes a protein with an immunoglobulin domain of unknown function, possibly involving interactions with the host immune system. It is similar in structure to the ORF7a protein, suggesting it may have originated through gene duplication.
ORF6 is a gene that encodes a viral accessory protein in coronaviruses of the subgenus Sarbecovirus, including SARS-CoV and SARS-CoV-2. It is not present in MERS-CoV. It is thought to reduce the immune system response to viral infection through interferon antagonism.

ORF9b is a gene that encodes a viral accessory protein in coronaviruses of the subgenus Sarbecovirus, including SARS-CoV and SARS-CoV-2. It is an overlapping gene whose open reading frame is entirely contained within the N gene, which encodes coronavirus nucleocapsid protein. The encoded protein is 97 amino acid residues long in SARS-CoV and 98 in SARS-CoV-2, in both cases forming a protein dimer.