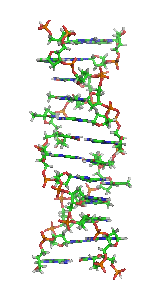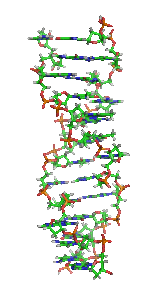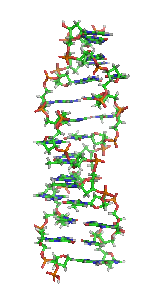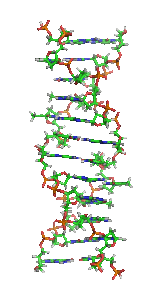
Z-DNA is one of the many possible double helical structures of DNA. It is a left-handed double helical structure in which the helix winds to the left in a zigzag pattern, instead of to the right, like the more common B-DNA form. Z-DNA is thought to be one of three biologically active double-helical structures along with A-DNA and B-DNA.

RNA editing is a molecular process through which some cells can make discrete changes to specific nucleotide sequences within an RNA molecule after it has been generated by RNA polymerase. It occurs in all living organisms and is one of the most evolutionarily conserved properties of RNAs. RNA editing may include the insertion, deletion, and base substitution of nucleotides within the RNA molecule. RNA editing is relatively rare, with common forms of RNA processing not usually considered as editing. It can affect the activity, localization as well as stability of RNAs, and has been linked with human diseases.

Potassium voltage-gated channel subfamily A member 1 also known as Kv1.1 is a shaker related voltage-gated potassium channel that in humans is encoded by the KCNA1 gene. The Isaacs syndrome is a result of an autoimmune reaction against the Kv1.1 ion channel.

Glutamate receptor 3 is a protein that in humans is encoded by the GRIA3 gene.

Filamin A, alpha (FLNA) is a protein that in humans is encoded by the FLNA gene.

Double-stranded RNA-specific adenosine deaminase is an enzyme that in humans is encoded by the ADAR gene.

Glutamate ionotropic receptor AMPA type subunit 2 is a protein that in humans is encoded by the GRIA2 gene and it is a subunit found in the AMPA receptors.

Insulin-like growth factor-binding protein 7 is a protein that in humans is encoded by the IGFBP7 gene. The major function of the protein is the regulation of availability of insulin-like growth factors (IGFs) in tissue as well as in modulating IGF binding to its receptors. IGFBP7 binds to IGF with high affinity. It also stimulates cell adhesion. The protein is implicated in some cancers.

Double-stranded RNA-specific editase 1 is an enzyme that in humans is encoded by the ADARB1 gene.

Glutamate receptor, ionotropic, kainate 1, also known as GRIK1, is a protein that in humans is encoded by the GRIK1 gene.

Cytoplasmic FMR1-interacting protein 2 is a protein that in humans is encoded by the CYFIP2 gene. Cytoplasmic FMR1 interacting protein is a 1253 amino acid long protein and is highly conserved sharing 99% sequence identity to the mouse protein. It is expressed mainly in brain tissues, white blood cells and the kidney.

Glutamate receptor 4 is a protein that in humans is encoded by the GRIA4 gene.

Gamma-aminobutyric acid receptor subunit alpha-3 is a protein that in humans is encoded by the GABRA3 gene.

Bladder cancer-associated protein is a protein that in humans is encoded by the BLCAP gene.

Double-stranded RNA-specific editase B2 is an enzyme that in humans is encoded by the ADARB2 gene.

ADP-ribosylation-like factor 6 interacting protein 4 (ARL6IP4), also called SRp25 is the product of the ARL6IP4 gene located on chromosome 12q24. 31. Its function is unknown.
In human genetics, the GRIA2 gene is located on chromosome 4q32-q33. The gene product is the ionotropic AMPA glutamate receptor 2. The protein belongs to a family of ligand-activated glutamate receptors that are sensitive to alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate (AMPA). Glutamate receptors function as the main excitatory neurotransmitter at many synapses in the central nervous system. L-glutamate, an excitatory neurotransmitter, binds to the Gria2 resulting in a conformational change. This leads to the opening of the channel converting the chemical signal to an electrical impulse. AMPA receptors (AMPAR) are composed of four subunits, designated as GluR1 (GRIA1), GluR2 (GRIA2), GluR3 (GRIA3), and GluR4(GRIA4) which combine to form tetramers. They are usually heterotrimeric but can be homodimeric. Each AMPAR has four sites to which an agonist can bind, one for each subunit.[5]

The complement component 1, q subcomponent-like 1 is encoded by a gene located at chromosome 17q21.31. It is a secreted protein and is 258 amino acids in length. The protein is widely expressed but its expression is highest in the brain and may also be involved in regulation of motor control. The pre-mRNA of this protein is subject to RNA editing.
Z-RNA is a left-handed alternative conformation for the RNA double helix. Just like for Z-DNA, Z-RNA is favored by a sequence composed of Purine/Pyrimidine repeats and especially CG repeats.

LEAPER is a genetic engineering technique in molecular biology by which RNA can be edited. The technique relies on engineered strands of RNA to recruit native ADAR enzymes to swap out different compounds in RNA. Developed by researchers at Peking University in 2019, the technique, some have claimed, is more efficient than the CRISPR gene editing technique. Initial studies have claimed that editing efiiciencies of up to 80% can be achieved.
















