
Methionine is an essential amino acid in humans.

In biochemistry, the DNA methyltransferase family of enzymes catalyze the transfer of a methyl group to DNA. DNA methylation serves a wide variety of biological functions. All the known DNA methyltransferases use S-adenosyl methionine (SAM) as the methyl donor.

Histone methyltransferases (HMT) are histone-modifying enzymes, that catalyze the transfer of one, two, or three methyl groups to lysine and arginine residues of histone proteins. The attachment of methyl groups occurs predominantly at specific lysine or arginine residues on histones H3 and H4. Two major types of histone methyltranferases exist, lysine-specific and arginine-specific. In both types of histone methyltransferases, S-Adenosyl methionine (SAM) serves as a cofactor and methyl donor group.
The genomic DNA of eukaryotes associates with histones to form chromatin. The level of chromatin compaction depends heavily on histone methylation and other post-translational modifications of histones. Histone methylation is a principal epigenetic modification of chromatin that determines gene expression, genomic stability, stem cell maturation, cell lineage development, genetic imprinting, DNA methylation, and cell mitosis.
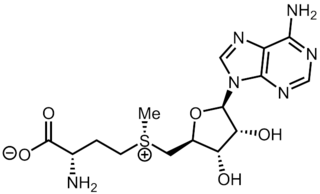
S-Adenosyl methionine (SAM), also known under the commercial names of SAMe, SAM-e, or AdoMet, is a common cosubstrate involved in methyl group transfers, transsulfuration, and aminopropylation. Although these anabolic reactions occur throughout the body, most SAM is produced and consumed in the liver. More than 40 methyl transfers from SAM are known, to various substrates such as nucleic acids, proteins, lipids and secondary metabolites. It is made from adenosine triphosphate (ATP) and methionine by methionine adenosyltransferase. SAM was first discovered by Giulio Cantoni in 1952.

The Structural Classification of Proteins (SCOP) database is a largely manual classification of protein structural domains based on similarities of their structures and amino acid sequences. A motivation for this classification is to determine the evolutionary relationship between proteins. Proteins with the same shapes but having little sequence or functional similarity are placed in different superfamilies, and are assumed to have only a very distant common ancestor. Proteins having the same shape and some similarity of sequence and/or function are placed in "families", and are assumed to have a closer common ancestor.

Methyltransferases are a large group of enzymes that all methylate their substrates but can be split into several subclasses based on their structural features. The most common class of methyltransferases is class I, all of which contain a Rossmann fold for binding S-Adenosyl methionine (SAM). Class II methyltransferases contain a SET domain, which are exemplified by SET domain histone methyltransferases, and class III methyltransferases, which are membrane associated. Methyltransferases can also be grouped as different types utilizing different substrates in methyl transfer reactions. These types include protein methyltransferases, DNA/RNA methyltransferases, natural product methyltransferases, and non-SAM dependent methyltransferases. SAM is the classical methyl donor for methyltransferases, however, examples of other methyl donors are seen in nature. The general mechanism for methyl transfer is a SN2-like nucleophilic attack where the methionine sulfur serves as the leaving group and the methyl group attached to it acts as the electrophile that transfers the methyl group to the enzyme substrate. SAM is converted to S-Adenosyl homocysteine (SAH) during this process. The breaking of the SAM-methyl bond and the formation of the substrate-methyl bond happen nearly simultaneously. These enzymatic reactions are found in many pathways and are implicated in genetic diseases, cancer, and metabolic diseases. Another type of methyl transfer is the radical S-Adenosyl methionine (SAM) which is the methylation of unactivated carbon atoms in primary metabolites, proteins, lipids, and RNA.

A leucine-rich repeat (LRR) is a protein structural motif that forms an α/β horseshoe fold. It is composed of repeating 20–30 amino acid stretches that are unusually rich in the hydrophobic amino acid leucine. These tandem repeats commonly fold together to form a solenoid protein domain, termed leucine-rich repeat domain. Typically, each repeat unit has beta strand-turn-alpha helix structure, and the assembled domain, composed of many such repeats, has a horseshoe shape with an interior parallel beta sheet and an exterior array of helices. One face of the beta sheet and one side of the helix array are exposed to solvent and are therefore dominated by hydrophilic residues. The region between the helices and sheets is the protein's hydrophobic core and is tightly sterically packed with leucine residues.

DNA adenine methylase, (Dam) (also site-specific DNA-methyltransferase (adenine-specific), EC 2.1.1.72, modification methylase, restriction-modification system) is an enzyme that adds a methyl group to the adenine of the sequence 5'-GATC-3' in newly synthesized DNA. Immediately after DNA synthesis, the daughter strand remains unmethylated for a short time. It is an orphan methyltransferase that is not part of a restriction-modification system and regulates gene expression. This enzyme catalyses the following chemical reaction

In molecular biology, a protein domain is a region of a protein's polypeptide chain that is self-stabilizing and that folds independently from the rest. Each domain forms a compact folded three-dimensional structure. Many proteins consist of several domains, and a domain may appear in a variety of different proteins. Molecular evolution uses domains as building blocks and these may be recombined in different arrangements to create proteins with different functions. In general, domains vary in length from between about 50 amino acids up to 250 amino acids in length. The shortest domains, such as zinc fingers, are stabilized by metal ions or disulfide bridges. Domains often form functional units, such as the calcium-binding EF hand domain of calmodulin. Because they are independently stable, domains can be "swapped" by genetic engineering between one protein and another to make chimeric proteins.

Cystathionine-β-synthase, also known as CBS, is an enzyme (EC 4.2.1.22) that in humans is encoded by the CBS gene. It catalyzes the first step of the transsulfuration pathway, from homocysteine to cystathionine:

Adenosylhomocysteinase (EC 3.13.2.1, S-adenosylhomocysteine synthase, S-adenosylhomocysteine hydrolase, adenosylhomocysteine hydrolase, S-adenosylhomocysteinase, SAHase, AdoHcyase) is an enzyme that catalyzes the nicotinamide adenine dinucleotide (NAD+) dependent, reversible hydrolysis of S-adenosylhomocysteine to homocysteine and adenosine.
The enzyme adenosylmethionine decarboxylase catalyzes the conversion of S-adenosyl methionine to S-adenosylmethioninamine. Polyamines such as spermidine and spermine are essential for cellular growth under most conditions, being implicated in many cellular processes including DNA, RNA and protein synthesis. S-adenosylmethionine decarboxylase (AdoMetDC) plays an essential regulatory role in the polyamine biosynthetic pathway by generating the n-propylamine residue required for the synthesis of spermidine and spermine from putrescein. Unlike many amino acid decarboxylases AdoMetDC uses a covalently bound pyruvate residue as a cofactor rather than the more common pyridoxal 5'-phosphate. These proteins can be divided into two main groups which show little sequence similarity either to each other, or to other pyruvoyl-dependent amino acid decarboxylases: class I enzymes found in bacteria and archaea, and class II enzymes found in eukaryotes. In both groups the active enzyme is generated by the post-translational autocatalytic cleavage of a precursor protein. This cleavage generates the pyruvate precursor from an internal serine residue and results in the formation of two non-identical subunits termed alpha and beta which form the active enzyme.

In enzymology, a protein-glutamate O-methyltransferase is an enzyme that catalyzes the chemical reaction
In molecular biology, the CBS domain is a protein domain found in a range of proteins in all species from bacteria to humans. It was first identified as a conserved sequence region in 1997 and named after cystathionine beta synthase, one of the proteins it is found in. CBS domains are also found in a wide variety of other proteins such as inosine monophosphate dehydrogenase, voltage gated chloride channels and AMP-activated protein kinase (AMPK). CBS domains regulate the activity of associated enzymatic and transporter domains in response to binding molecules with adenosyl groups such as AMP and ATP, or s-adenosylmethionine.

S-Adenosylmethionine synthetase, also known as methionine adenosyltransferase (MAT), is an enzyme that creates S-adenosylmethionine by reacting methionine and ATP.

The SET domain is a protein domain that typically has methyltransferase activity. It was originally identified as part of a larger conserved region present in the Drosophila Trithorax protein and was subsequently identified in the Drosophila Su(var)3-9 and 'Enhancer of zeste' proteins, from which the acronym SET is derived [Su(var)3-9, Enhancer-of-zeste and Trithorax].
Radical SAM enzymes belong to a superfamily of enzymes that use an iron-sulfur cluster (4Fe-4S) to reductively cleave S-adenosyl-L-methionine (SAM) to generate a radical, usually a 5′-deoxyadenosyl radical (5'-dAdo), as a critical intermediate. These enzymes utilize this radical intermediate to perform diverse transformations, often to functionalize unactivated C-H bonds. Radical SAM enzymes are involved in cofactor biosynthesis, enzyme activation, peptide modification, post-transcriptional and post-translational modifications, metalloprotein cluster formation, tRNA modification, lipid metabolism, biosynthesis of antibiotics and natural products etc. The vast majority of known radical SAM enzymes belong to the radical SAM superfamily, and have a cysteine-rich motif that matches or resembles CxxxCxxC. Radical SAM enzymes comprise the largest superfamily of metal-containing enzymes.
23S rRNA (adenine2503-C8)-methyltransferase (EC 2.1.1.224, Cfr (gene)) is an enzyme with systematic name S-adenosyl-L-methionine:23S rRNA (adenine2503-C8)-methyltransferase. This enzyme catalyses the following chemical reaction
Protein methylation is a type of post-translational modification featuring the addition of methyl groups to proteins. It can occur on the nitrogen-containing side-chains of arginine and lysine, but also at the amino- and carboxy-termini of a number of different proteins. In biology, methyltransferases catalyze the methylation process, activated primarily by S-adenosylmethionine. Protein methylation has been most studied in histones, where the transfer of methyl groups from S-adenosyl methionine is catalyzed by histone methyltransferases. Histones that are methylated on certain residues can act epigenetically to repress or activate gene expression.
Methylthiotransferases are enzymes of the radical S-adenosyl methionine superfamily. These enzymes catalyze the addition of a methylthio group to various biochemical compounds including tRNA and proteins. Methylthiotransferases are classified into one of four classes based on their substrates and mechanisms. All methylthiotransferases have been shown to contain two Fe-S clusters, one canonical cluster and one auxiliary cluster, that both function in the addition of the methylthio group to the substrate.
















