
The human microbiome is the aggregate of all microbiota that reside on or within human tissues and biofluids along with the corresponding anatomical sites in which they reside, including the skin, mammary glands, seminal fluid, uterus, ovarian follicles, lung, saliva, oral mucosa, conjunctiva, biliary tract, and gastrointestinal tract. Types of human microbiota include bacteria, archaea, fungi, protists, and viruses. Though micro-animals can also live on the human body, they are typically excluded from this definition. In the context of genomics, the term human microbiome is sometimes used to refer to the collective genomes of resident microorganisms; however, the term human metagenome has the same meaning.
In medicine, the hygiene hypothesis states that early childhood exposure to particular microorganisms protects against allergies by strengthening the immune system. In particular, a lack of such exposure is thought to lead to poor immune tolerance. The time period for exposure begins before birth and ends at school age.

Gut microbiota, gut microbiome, or gut flora, are the microorganisms, including bacteria, archaea, fungi, and viruses, that live in the digestive tracts of animals. The gastrointestinal metagenome is the aggregate of all the genomes of the gut microbiota. The gut is the main location of the human microbiome. The gut microbiota has broad impacts, including effects on colonization, resistance to pathogens, maintaining the intestinal epithelium, metabolizing dietary and pharmaceutical compounds, controlling immune function, and even behavior through the gut–brain axis.

Bacteroides is a genus of Gram-negative, obligate anaerobic bacteria. Bacteroides species are non endospore-forming bacilli, and may be either motile or nonmotile, depending on the species. The DNA base composition is 40–48% GC. Unusual in bacterial organisms, Bacteroides membranes contain sphingolipids. They also contain meso-diaminopimelic acid in their peptidoglycan layer.

Bacteroides fragilis is an anaerobic, Gram-negative, pleomorphic to rod-shaped bacterium. It is part of the normal microbiota of the human colon and is generally commensal, but can cause infection if displaced into the bloodstream or surrounding tissue following surgery, disease, or trauma.
Dysbiosis is characterized by a disruption to the microbiome resulting in an imbalance in the microbiota, changes in their functional composition and metabolic activities, or a shift in their local distribution. For example, a part of the human microbiota such as the skin flora, gut flora, or vaginal flora, can become deranged, with normally dominating species underrepresented and normally outcompeted or contained species increasing to fill the void. Dysbiosis is most commonly reported as a condition in the gastrointestinal tract.

Long-term close-knit interactions between symbiotic microbes and their host can alter host immune system responses to other microorganisms, including pathogens, and are required to maintain proper homeostasis. The immune system is a host defense system consisting of anatomical physical barriers as well as physiological and cellular responses, which protect the host against harmful microorganisms while limiting host responses to harmless symbionts. Humans are home to 1013 to 1014 bacteria, roughly equivalent to the number of human cells, and while these bacteria can be pathogenic to their host most of them are mutually beneficial to both the host and bacteria.

Microbiota are the range of microorganisms that may be commensal, mutualistic, or pathogenic found in and on all multicellular organisms, including plants. Microbiota include bacteria, archaea, protists, fungi, and viruses, and have been found to be crucial for immunologic, hormonal, and metabolic homeostasis of their host.

Porphyromonas is a Gram-negative, non-spore-forming, obligately anaerobic and non-motile genus from the family Porphyromonadaceae. There were 16 different Porphyromonas species documented as of 2015, which reside in both animal and human reservoirs. It was discovered more recently that Porphyromonas also exist in the environment, albeit to a lesser extent. This genus is notably implicated in the modulation of oral cavity, respiratory tract, and gastrointestinal tract disease states. It is suggested that Porphyromonas either operate as benign bacteria pertinent to host immunity or are potential pathobionts that opportunistically provoke diseased states when homeostasis is disrupted. Despite its characterization not being fully elucidated due to sparse research, various studies report the prevalence of this genus at 58.7% in healthy states compared with 41.3% in diseased states.
Metaproteomics is an umbrella term for experimental approaches to study all proteins in microbial communities and microbiomes from environmental sources. Metaproteomics is used to classify experiments that deal with all proteins identified and quantified from complex microbial communities. Metaproteomics approaches are comparable to gene-centric environmental genomics, or metagenomics.
An enterotype is a classification of living organisms based on the bacteriological composition of their gut microbiota. The discovery of three human enterotypes was announced in the April 2011 issue of Nature by Peer Bork and his associates. They found that enterotypes are not dictated by age, gender, body weight, or national divisions. There are indications that long-term diet influences enterotype. Type 1 is characterized by high levels of Bacteroides, type 2 has few Bacteroides but Prevotella are common, and type 3 has high levels of Ruminococcus.
The altered Schaedler flora (ASF) is a community of eight bacterial species: two lactobacilli, one Bacteroides, one spiral bacterium of the Flexistipes genus, and four extremely oxygen sensitive (EOS) fusiform-shaped species. The bacteria are selected for their dominance and persistence in the normal microflora of mice, and for their ability to be isolated and grown in laboratory settings. Germ-free animals, mainly mice, are colonized with ASF for the purpose of studying the gastrointestinal (GI) tract. Intestinal mutualistic bacteria play an important role in affecting gene expression of the GI tract, immune responses, nutrient absorption, and pathogen resistance. The standardized microbial cocktail enabled the controlled study of microbe and host interactions, role of microbes, pathogen effects, and intestinal immunity and disease association, such as cancer, inflammatory bowel disease, diabetes, and other inflammatory or autoimmune diseases. Also, compared to germfree animals, ASF mice have fully developed immune system, resistance to opportunistic pathogens, and normal GI function and health, and are a great representation of normal mice.
Sarkis Mazmanian is an American medical microbiologist who has served as a professor at the California Institute of Technology since 2006. He is currently the Luis & Nelly Soux Professor of Microbiology in the Division of Biology and Biological Engineering, and a board member of Seed. Prior to this, Mazmanian was affiliated with Harvard Medical School and the University of Chicago. In 2012, Mazmanian was awarded a MacArthur Fellowship for his pioneering work on the human microbiome.
The microbiota are the sum of all symbiotic microorganisms living on or in an organism. The fruit fly Drosophila melanogaster is a model organism and known as one of the most investigated organisms worldwide. The microbiota in flies is less complex than that found in humans. It still has an influence on the fitness of the fly, and it affects different life-history characteristics such as lifespan, resistance against pathogens (immunity) and metabolic processes (digestion). Considering the comprehensive toolkit available for research in Drosophila, analysis of its microbiome could enhance our understanding of similar processes in other types of host-microbiota interactions, including those involving humans. Microbiota plays key roles in the intestinal immune and metabolic responses via their fermentation product, acetate.
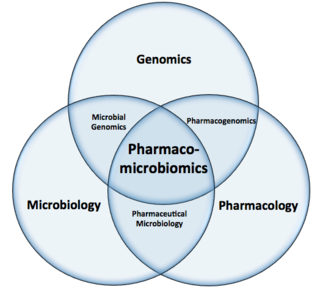
Pharmacomicrobiomics, proposed by Prof. Marco Candela for the ERC-2009-StG project call, and publicly coined for the first time in 2010 by Rizkallah et al., is defined as the effect of microbiome variations on drug disposition, action, and toxicity. Pharmacomicrobiomics is concerned with the interaction between xenobiotics, or foreign compounds, and the gut microbiome. It is estimated that over 100 trillion prokaryotes representing more than 1000 species reside in the gut. Within the gut, microbes help modulate developmental, immunological and nutrition host functions. The aggregate genome of microbes extends the metabolic capabilities of humans, allowing them to capture nutrients from diverse sources. Namely, through the secretion of enzymes that assist in the metabolism of chemicals foreign to the body, modification of liver and intestinal enzymes, and modulation of the expression of human metabolic genes, microbes can significantly impact the ingestion of xenobiotics.
The human milk microbiota, also known as human milk probiotics (HMP), refers to the microbiota (community of microorganisms) residing in the human mammary glands and breast milk. Human breast milk has been traditionally assumed to be sterile, but more recently both microbial culture and culture-independent techniques have confirmed that human milk contains diverse communities of bacteria which are distinct from other microbial communities inhabiting the human body.

Bacteroides thetaiotaomicron is a gram-negative, rod shaped obligate anaerobic bacterium that is a prominent member of the normal gut microbiome in the distal intestines. Its proteome, consisting of 4,779 members, includes a system for obtaining and breaking down dietary polysaccharides that would otherwise be difficult to digest. B. thetaiotaomicron is also an opportunistic pathogen, meaning it may become virulent in immunocompromised individuals. It is often used in research as a model organism for functional studies of the human microbiota.

Abigail A. Salyers was a microbiologist who pioneered the field of human microbiome research. Her work on the bacterial phylum Bacteroidetes and its ecology led to a better understanding of antibiotic resistance and mobile genetic elements. At a time where the prevailing paradigm was focused on E. coli as a model organism, Salyers emphasized the importance of investigating the breadth of microbial diversity. She was one of the first to conceptualize the human body as a microbial ecosystem. Over the course of her 40-year career, she was presented with numerous awards for teaching and research and an honorary degree from ETH Zurich, and served as president of the American Society for Microbiology.

The cetacean microbiome is the group of communities of microorganisms that reside within whales.

Phocaeicola vulgatus,, is a mutualistic anaerobic Gram negative rod bacteria commonly found in the human gut microbiome and isolated from feces. P. vulgatus has medical relevance and has been notable in scientific research due to its production of fatty acids, potential use as a probiotic, and associations with protecting against and worsening some inflammatory diseases. Due to the difficulties in culturing anaerobic bacteria, P. vulgatus is still highly uncharacterised so efforts are being made to make use of multi-omic approaches to investigate the human gut microbiome more thoroughly in hopes to fully understand the role of this species in the development of and protection against diseases, as well as its potential uses in medicine and research. Generally, P. vulgatus is considered as a beneficial bacteria that contributes to digestion and a balanced microbiome, but it has been known to cause opportunistic infections and induce or worsen inflammatory responses. Due to its abundance in the microbiome, some researchers are investigating these species in hopes that it will be a suitable model organism for gut microbiome research, like Bacteroides thetaiotaomicron.











