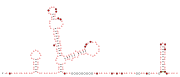
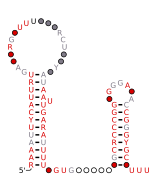DsrA RNA is a non-coding RNA that regulates both transcription, by overcoming transcriptional silencing by the nucleoid-associated H-NS protein, and translation, by promoting efficient translation of the stress sigma factor, RpoS. These two activities of DsrA can be separated by mutation: the first of three stem-loops of the 85 nucleotide RNA is necessary for RpoS translation but not for anti-H-NS action, while the second stem-loop is essential for antisilencing and less critical for RpoS translation. The third stem-loop, which behaves as a transcription terminator, can be substituted by the trp transcription terminator without loss of either DsrA function. The sequence of the first stem-loop of DsrA is complementary with the upstream leader portion of RpoS messenger RNA, suggesting that pairing of DsrA with the RpoS message might be important for translational regulation. The structures of DsrA and DsrA/rpoS complex were studied by NMR. The study concluded that the sRNA contains a dynamic conformational equilibrium for its second stem–loop which might be an important mechanism for DsrA to regulate the translations of its multiple target mRNAs.
RNAIII is a stable 514 nt regulatory RNA transcribed by the P3 promoter of the Staphylococcus aureus quorum-sensing agr system ). It is the major effector of the agr regulon, which controls the expression of many S. aureus genes encoding exoproteins and cell wall associated proteins plus others encoding regulatory proteins The RNAIII transcript also encodes the 26 amino acid δ-haemolysin peptide (Hld). RNAIII contains many stem loops, most of which match the Shine-Dalgarno sequence involved in translation initiation of the regulated genes. Some of these interactions are inhibitory, others stimulatory; among the former is the regulatory protein Rot. In vitro, RNAIII is expressed post exponentially, inhibiting translation of the surface proteins, notably protein A, while stimulating that of the exoproteins, many of which are tissue-degrading enzymes or cytolysins. Among the latter is the important virulence factor, α-hemolysin (Hla), whose translation RNAIII activates by preventing the formation of an inhibitory foldback loop in the hla mRNA leader.
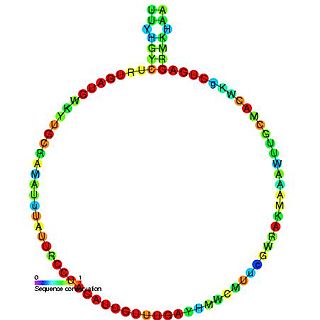
In molecular biology, snoRNA U34 is a non-coding RNA (ncRNA) molecule which functions in the modification of other small nuclear RNAs (snRNAs). This type of modifying RNA is usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA.

Spot 42 (spf) RNA is a regulatory non-coding bacterial small RNA encoded by the spf gene. Spf is found in gammaproteobacteria and the majority of experimental work on Spot42 has been performed in Escherichia coli and recently in Aliivibrio salmonicida. In the cell Spot42 plays essential roles as a regulator in carbohydrate metabolism and uptake, and its expression is activated by glucose, and inhibited by the cAMP-CRP complex.

The MicA RNA is a small non-coding RNA that was discovered in E. coli during a large scale screen. Expression of SraD is highly abundant in stationary phase, but low levels could be detected in exponentially growing cells as well.

The Hfq protein encoded by the hfq gene was discovered in 1968 as an Escherichia coli host factor that was essential for replication of the bacteriophage Qβ. It is now clear that Hfq is an abundant bacterial RNA binding protein which has many important physiological roles that are usually mediated by interacting with Hfq binding sRNA.
Rhodobacter sphaeroides is a kind of purple bacterium; a group of bacteria that can obtain energy through photosynthesis. Its best growth conditions are anaerobic phototrophy and aerobic chemoheterotrophy in the absence of light. R. sphaeroides is also able to fix nitrogen. It is remarkably metabolically diverse, as it is able to grow heterotrophically via fermentation and aerobic and anaerobic respiration. Such a metabolic versatility has motivated the investigation of R. sphaeroides as a microbial cell factory for biotechnological applications.

NrrF is a non-coding RNA which is regulated by the Ferric uptake regulator (Fur) protein in bacteria. This non-coding RNA was identified in Neisseria meningitidis and is involved in iron regulation of the succinate dehydrogenase genes sdhA and sdhC. NrrF acts as an antisense RNA and is complementary to the junction between the second and third genes of the sdh operon. Secondary structure predictions have indicated that this interaction occurs in a single stranded loop region of the NrrF RNA. Under low iron concentration NrrF is present at a high concentration and forms a duplex with the transcript in Hfq dependent manner. The RNA chaperone Hfq acts to enhance binding of NrrF or stabilizes the NrrF/sdh transcript duplex. Binding of NrrF results in down regulation of the sdhCDAB mRNA transcript results in a Fur-dependent positive regulation of succinate dehydrogenase. Another NrrF RNA target is mRNA petABC, coding for cytochrome bc1. Interaction between NrrF and the 5′ untranslated region of the petABC mRNA results in its repression.

The IscR stability element is a conserved secondary structure found in the intergenic regions of iscRSUA polycistronic mRNA. This secondary structure prevents the degradation of the iscR mRNA.
Pseudomonas sRNA are non-coding RNAs (ncRNA) that were predicted by the bioinformatic program SRNApredict2. This program identifies putative sRNAs by searching for co-localization of genetic features commonly associated with sRNA-encoding genes and the expression of the predicted sRNAs was subsequently confirmed by Northern blot analysis. These sRNAs have been shown to be conserved across several pseudomonas species but their function is yet to be determined. Using Tet-Trap genetic approach RNAT genes post-transcriptionally regulated by temperature upshift were identified: ptxS and PA5194.

The Bacteroidales-1 RNA motif is a conserved RNA structure identified by bioinformatics. It has been identified only in bacteria within the order (biology) Bacteroidales. Its presumed length is marked by a promoter on one end that conforms to an alternate consensus sequence that is common in the phylum Bacteroidota, and its 3′ end is indicated by predicted transcription terminators. It is often located downstream of a gene that encodes the L20 ribosomal subunit, although it is unclear whether there is a functional reason underlying this apparent association.
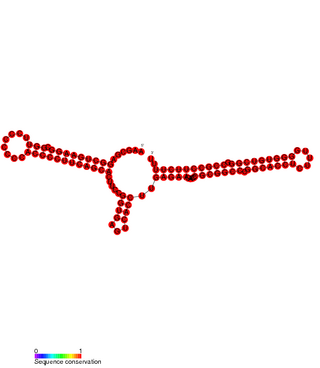
Pxr sRNA is a regulatory RNA which downregulates genes responsible for the formation of fruiting bodies in Myxococcus xanthus. Fruiting bodies are aggregations of myxobacteria formed when nutrients are scarce, the fruiting bodies permit a small number of the aggregated colony to transform into stress-resistant spores.

A toxin-antitoxin system consists of a "toxin" and a corresponding "antitoxin", usually encoded by closely linked genes. The toxin is usually a protein while the antitoxin can be a protein or an RNA. Toxin-antitoxin systems are widely distributed in prokaryotes, and organisms often have them in multiple copies. When these systems are contained on plasmids – transferable genetic elements – they ensure that only the daughter cells that inherit the plasmid survive after cell division. If the plasmid is absent in a daughter cell, the unstable antitoxin is degraded and the stable toxic protein kills the new cell; this is known as 'post-segregational killing' (PSK).
Rsa RNAs are non-coding RNAs found in the bacterium Staphylococcus aureus. The shared name comes from their discovery, and does not imply homology. Bioinformatics scans identified the 16 Rsa RNA families named RsaA-K and RsaOA-OG. Others, RsaOH-OX, were found thanks to an RNomic approach. Although the RNAs showed varying expression patterns, many of the newly discovered RNAs were shown to be Hfq-independent and most carried a C-rich motif (UCCC).
In molecular biology, 5' ureB sRNA is a small RNA. It is located at the 5' end of the ureB gene in the urease gene cluster and is antisense to ureB. Its expression is regulated by the arsS/arsR two-component regulatory system, arsS downregulates 5' ureB sRNA expression under acidic conditions leading to an increase in ureB expression. 5' ureB sRNA suppresses transcription of ureA and ureB by base-pairing with ureAB mRNA resulting in premature termination of the transcript to the 5' of ureB.
The branched chain amino acid:cation symporter (LIVCS) family (TC# 2.A.26) is a member of the APC superfamily. Characterized members of this family transport all three of the branched chain aliphatic amino acids (leucine (L), isoleucine (I) and valine (V)). These proteins are found in Gram-negative and Gram-positive bacteria and function by a Na+ or H+ symport mechanism. They possess about 440 amino acyl residues and display 12 putative transmembrane helical spanners. As of early 2016, no crystal structures for members of the LIVCS family are available on RCSB.
The NhaC family belongs to the Ion Transporter (IT) Superfamily. A representative list of proteins belonging to the NhaC family can be found in the Transporter Classification Database.
The CidA/LrgA Holin Family is a group of proteins named after CidA and LrgA of Staphylococcus aureus. CidA and LrgA are homologous holin and anti-holin proteins, each with 4 putative transmembrane segments (TMSs). Members of the CidA/LrgA holin family also include putative murine hydrolase exporters from a wide range of Gram-positive and Gram-negative bacteria as well as archaea. Most CidA/LrgA holin family proteins vary in size between 100 and 160 amino acyl residues (aas) in length although a few are larger.

Bacteroides thetaiotaomicron is a gram-negative, rod shaped obligate anaerobic bacterium that is a prominent member of the normal gut microbiome in the distal intestines. Its proteome, consisting of 4,779 members, includes a system for obtaining and breaking down dietary polysaccharides that would otherwise be difficult to digest. B. thetaiotaomicron is also an opportunistic pathogen, meaning it may become virulent in immunocompromised individuals. It is often used in research as a model organism for functional studies of the human microbiota.
Accessory gene regulator (agr) is a complex 5 gene locus that is a global regulator of virulence in Staphylococcus aureus. It encodes a two-component transcriptional quorum-sensing (QS) system activated by an autoinducing, thiolactone-containing cyclic peptide (AIP).