Related Research Articles

Functional genomics is a field of molecular biology that attempts to describe gene functions and interactions. Functional genomics make use of the vast data generated by genomic and transcriptomic projects. Functional genomics focuses on the dynamic aspects such as gene transcription, translation, regulation of gene expression and protein–protein interactions, as opposed to the static aspects of the genomic information such as DNA sequence or structures. A key characteristic of functional genomics studies is their genome-wide approach to these questions, generally involving high-throughput methods rather than a more traditional "candidate-gene" approach.

Pseudomonas aeruginosa is a common encapsulated, Gram-negative, aerobic–facultatively anaerobic, rod-shaped bacterium that can cause disease in plants and animals, including humans. A species of considerable medical importance, P. aeruginosa is a multidrug resistant pathogen recognized for its ubiquity, its intrinsically advanced antibiotic resistance mechanisms, and its association with serious illnesses – hospital-acquired infections such as ventilator-associated pneumonia and various sepsis syndromes.
Pseudomonas citronellolis is a Gram-negative, bacillus bacterium that is used to study the mechanisms of pyruvate carboxylase. It was first isolated from forest soil, under pine trees, in northern Virginia, United States.

The PrrF RNAs are small non-coding RNAs involved in iron homeostasis and are encoded by all Pseudomonas species. The PrrF RNAs are analogs of the RyhB RNA, which is encoded by enteric bacteria. Expression of the PrrF RNAs is repressed by the ferric uptake regulator (Fur) when cells are grown in iron-replete conditions. Under iron limitation, the PrrF RNAs are expressed and act to negatively regulate several genes encoding iron-containing proteins, including SodB and succinate dehydrogenase. As such, PrrF regulation "spares" iron when this nutrient becomes scarce.

Pseudomonas sRNA P1 is a ncRNA that was predicted using bioinformatic tools in the genome of the opportunistic pathogen Pseudomonas aeruginosa and its expression verified by northern blot analysis.
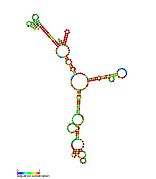
Pseudomonas sRNA P11 is a ncRNA that was predicted using bioinformatic tools in the genome of the opportunistic pathogen Pseudomonas aeruginosa and its expression verified by northern blot analysis. P11 is located between a putative threonine protein kinase and putative nitrate reductase and is conserved in several Pseudomonas species. P11 has a predicted Rho independent terminator at the 3′ end but the function of P11 is unknown.
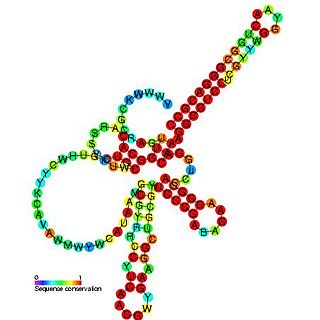
Pseudomonas sRNA P15 is a ncRNA that was predicted using bioinformatic tools in the genome of the opportunistic pathogen Pseudomonas aeruginosa and its expression verified by northern blot analysis.
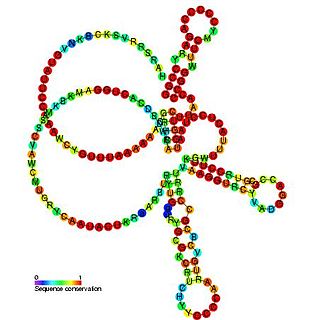
Pseudomonas sRNA P16 is a ncRNA that was predicted using bioinformatic tools in the genome of the opportunistic pathogen Pseudomonas aeruginosa and its expression verified by northern blot analysis. P16 sRNA appears to be conserved across several Pseudomonas species and is consistently located downstream of a predicted TatD deoxyribonuclease gene. P16 has a predicted Rho independent terminator at the 3′ end but the function of P16 is unknown.
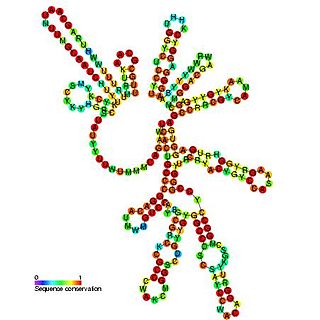
Pseudomonas sRNA P24 is a ncRNA that was predicted using bioinformatic tools in the genome of the opportunistic pathogen Pseudomonas aeruginosa and its expression verified by northern blot analysis.
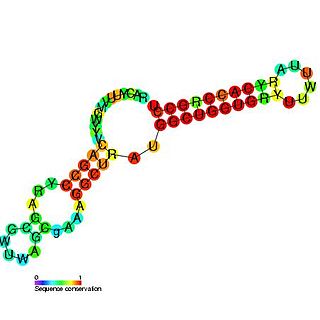
Pseudomonas sRNA P26 is a ncRNA that was predicted using bioinformatic tools in the genome of the opportunistic pathogen Pseudomonas aeruginosa and its expression verified by northern blot analysis. P26 is conserved across many Gammaproteobacteria species and appears to be consistently located between the DNA directed RNA polymerase and 50S ribosomal protein L7/L12 genes.

Pseudomonas sRNA P9 is a ncRNA that was predicted using bioinformatic tools in the genome of the opportunistic pathogen Pseudomonas aeruginosa and its expression verified by northern blot analysis.

RNA-Seq is a technique that uses next-generation sequencing (NGS) to reveal the presence and quantity of RNA molecules in a biological sample, providing a snapshot of gene expression in the sample, also known as transcriptome.
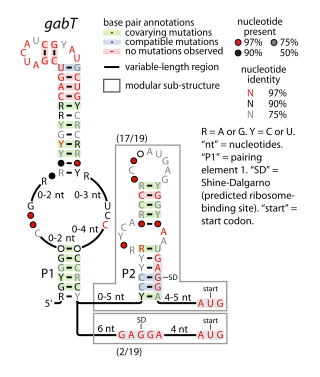
The gabT RNA motif is the name of a conserved RNA structure identified by bioinformatics whose function is unknown. The gabT motif has been detected exclusively in bacteria within the genus Pseudomonas, and is found only upstream of gabT genes, and downstream to gabD genes.

The gyrA RNA motif is a conserved RNA structure identified by bioinformatics. The RNAs are present in multiple species of bacteria within the order Pseudomonadales. This order contains the genus Pseudomonas, which includes the opportunistic human pathogen Pseudomonas aeruginosa and Pseudomonas syringae, a plant pathogen.

The Pseudomon-1 RNA motif is a conserved RNA identified by bioinformatics. It is used by most species whose genomes have been sequenced and that are classified within the genus Pseudomonas, and is also present in Azotobacter vinelandii, a closely related species. It is presumed to function as a non-coding RNA. Pseudomon-1 RNAs consistently have a downstream rho-independent transcription terminator.
Bacterial small RNAs (bsRNA) are small RNAs produced by bacteria; they are 50- to 500-nucleotide non-coding RNA molecules, highly structured and containing several stem-loops. Numerous sRNAs have been identified using both computational analysis and laboratory-based techniques such as Northern blotting, microarrays and RNA-Seq in a number of bacterial species including Escherichia coli, the model pathogen Salmonella, the nitrogen-fixing alphaproteobacterium Sinorhizobium meliloti, marine cyanobacteria, Francisella tularensis, Streptococcus pyogenes, the pathogen Staphylococcus aureus, and the plant pathogen Xanthomonas oryzae pathovar oryzae. Bacterial sRNAs affect how genes are expressed within bacterial cells via interaction with mRNA or protein, and thus can affect a variety of bacterial functions like metabolism, virulence, environmental stress response, and structure.

Mycobacterium tuberculosis contains at least nine small RNA families in its genome. The small RNA (sRNA) families were identified through RNomics – the direct analysis of RNA molecules isolated from cultures of Mycobacterium tuberculosis. The sRNAs were characterised through RACE mapping and Northern blot experiments. Secondary structures of the sRNAs were predicted using Mfold.

In molecular biology and genetics, DNA annotation or genome annotation is the process of describing the structure and function of the components of a genome, by analyzing and interpreting them in order to extract their biological significance and understand the biological processes in which they participate. Among other things, it identifies the locations of genes and all the coding regions in a genome and determines what those genes do.

Escherichia coli contains a number of small RNAs located in intergenic regions of its genome. The presence of at least 55 of these has been verified experimentally. 275 potential sRNA-encoding loci were identified computationally using the QRNA program. These loci will include false positives, so the number of sRNA genes in E. coli is likely to be less than 275. A computational screen based on promoter sequences recognised by the sigma factor sigma 70 and on Rho-independent terminators predicted 24 putative sRNA genes, 14 of these were verified experimentally by northern blotting. The experimentally verified sRNAs included the well characterised sRNAs RprA and RyhB. Many of the sRNAs identified in this screen, including RprA, RyhB, SraB and SraL, are only expressed in the stationary phase of bacterial cell growth. A screen for sRNA genes based on homology to Salmonella and Klebsiella identified 59 candidate sRNA genes. From this set of candidate genes, microarray analysis and northern blotting confirmed the existence of 17 previously undescribed sRNAs, many of which bind to the chaperone protein Hfq and regulate the translation of RpoS. UptR sRNA transcribed from the uptR gene is implicated in suppressing extracytoplasmic toxicity by reducing the amount of membrane-bound toxic hybrid protein.

The ivy-DE RNA motif is a conserved RNA structure that was discovered by bioinformatics. ivy-DE motifs are found in the genus Pseudomonas.
References
- ↑ Livny, J.; Brencic, A.; Lory, S.; Waldor, K. (2006). "Identification of 17 Pseudomonas aeruginosa sRNAs and prediction of sRNA-encoding genes in 10 diverse pathogens using the bioinformatic tool sRNAPredict2" (Free full text). Nucleic Acids Research. 34 (12): 3484–3493. doi:10.1093/nar/gkl453. ISSN 0305-1048. PMC 1524904 . PMID 16870723.
- ↑ Delvillani, Francesco; Sciandrone, Barbara; Peano, Clelia; Petiti, Luca; Berens, Christian; Georgi, Christiane; Ferrara, Silvia; Bertoni, Giovanni; Pasini, Maria Enrica (December 2014). "Tet-Trap, a genetic approach to the identification of bacterial RNA thermometers: application to Pseudomonas aeruginosa". RNA. 20 (12): 1963–1976. doi:10.1261/rna.044354.114. ISSN 1469-9001. PMC 4238360 . PMID 25336583.