
The Pseudomonadaceae are a family of bacteria which includes the genera Azomonas, Azorhizophilus, Azotobacter, Mesophilobacter, Pseudomonas, and Rugamonas. The family Azotobacteraceae was recently reclassified into this family.

The PrrF RNAs are small non-coding RNAs involved in iron homeostasis and are encoded by all Pseudomonas species. The PrrF RNAs are analogs of the RyhB RNA, which is encoded by enteric bacteria. Expression of the PrrF RNAs is repressed by the ferric uptake regulator (Fur) when cells are grown in iron-replete conditions. Under iron limitation, the PrrF RNAs are expressed and act to negatively regulate several genes encoding iron-containing proteins, including SodB and succinate dehydrogenase. As such, PrrF regulation "spares" iron when this nutrient becomes scarce.

The rsmY RNA family is a set of related non-coding RNA genes, that like RsmZ, is regulated by the GacS/GacA signal transduction system in the plant-beneficial soil bacterium and biocontrol model organism Pseudomonas fluorescens CHA0. GacA/GacS target genes are translationally repressed by the small RNA binding protein RsmA. RsmY and RsmZ RNAs bind RsmA to relieve this repression and so enhance secondary metabolism and biocontrol traits.

Swarming motility is a rapid and coordinated translocation of a bacterial population across solid or semi-solid surfaces, and is an example of bacterial multicellularity and swarm behaviour. Swarming motility was first reported by Jorgen Henrichsen and has been mostly studied in genus Serratia, Salmonella, Aeromonas, Bacillus, Yersinia, Pseudomonas, Proteus, Vibrio and Escherichia.
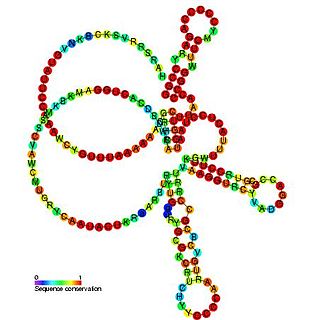
Pseudomonas sRNA P16 is a ncRNA that was predicted using bioinformatic tools in the genome of the opportunistic pathogen Pseudomonas aeruginosa and its expression verified by northern blot analysis. P16 sRNA appears to be conserved across several Pseudomonas species and is consistently located downstream of a predicted TatD deoxyribonuclease gene. P16 has a predicted Rho independent terminator at the 3'end but the function of P16 is unknown.
Pseudomonas sRNA are non-coding RNAs (ncRNA) that were predicted by the bioinformatic program SRNApredict2. This program identifies putative sRNAs by searching for co-localization of genetic features commonly associated with sRNA-encoding genes and the expression of the predicted sRNAs was subsequently confirmed by Northern blot analysis. These sRNAs have been shown to be conserved across several pseudomonas species but their function is yet to be determined. Using Tet-Trap genetic approach RNAT genes post-transcriptionally regulated by temperature upshift were identified: ptxS and PA5194.

The gabT RNA motif is the name of a conserved RNA structure identified by bioinformatics whose function is unknown. The gabT motif has been detected exclusively in bacteria within the genus Pseudomonas, and is found only upstream of gabT genes, and downstream to gabD genes.

The Pseudomon-1 RNA motif is a conserved RNA identified by bioinformatics. It is used by most species whose genomes have been sequenced and that are classified within the genus Pseudomonas, and is also present in Azotobacter vinelandii, a closely related species. It is presumed to function as a non-coding RNA. Pseudomon-1 RNAs consistently have a downstream rho-independent transcription terminator.
The Pseudomon-groES RNA motif is a conserved RNA structure identified in certain bacteria using bioinformatics. It is found in most species within the family Pseudomonadaceae, and is consistently located in the 5' untranslated regions of operons that contain groES genes. RNA transcripts of the groES genes in Pseudomonas aeruginosa where shown experimentally to be initiated at one of two start sites, from promoters called "P1" and "P2". The Pseudomon-groES RNA is in the 5' UTR of transcripts initiated from the P1 site, but is truncated in P2 transcripts. groES genes are involved in the cellular response to heat shock, but it is not thought that the Pseudomonas-groES RNA motif is involved in heat shock regulation. However, it is thought that the motif might regulate groES genes in response to other stimuli.
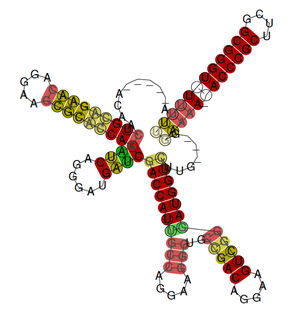
The rsmX gene is part of the Rsm/Csr family of non-coding RNAs (ncRNAs). Members of the Rsm/Csr family are present in a diverse range of bacteria, including Escherichia coli, Erwinia, Salmonella, Vibrio and Pseudomonas. These ncRNAs act by sequestering translational repressor proteins, called RsmA, activating expression of downstream genes that would normally be blocked by the repressors. Sequestering of target proteins is dependent upon exposed GGA motifs in the stem loops of the ncRNAs. Typically, the activated genes are involved in secondary metabolism, biofilm formation and motility.

Rhamnolipids are a class of glycolipid produced by Pseudomonas aeruginosa, amongst other organisms, frequently cited as the best characterised of the bacterial surfactants. They have a glycosyl head group, in this case a rhamnose moiety, and a 3-(hydroxyalkanoyloxy)alkanoic acid (HAA) fatty acid tail, such as 3-hydroxydecanoic acid.

CrcZ is a small RNA found in Pseudomonas bacteria, which acts as a global regulator of carbon catabolite repression. In P. aeruginosa, CrcZ is responsible for sequestering the protein Crc. Crc is an RNA-binding global regulator, which acts by inhibiting the translation of the transcriptional regulator AlkS.
PhrS is a bacterial small RNA found in Pseudomonas aeruginosa. It was first identified in a RNAomics screen and has since been found to act as a link between oxygen availability and quorum sensing.
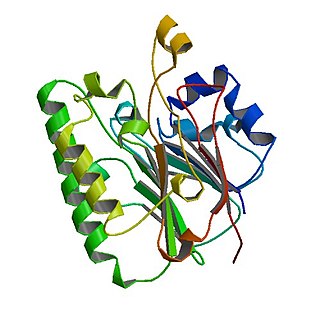
The Catabolite repression control (Crc) protein participates in suppressing expression of several genes involved in utilization of carbon sources in Pseudomonas bacteria. Presence of organic acids triggers activation of Crc and in conjunction with the Hfq protein genes that metabolize a given carbon source are downregulated until another more favorable carbon source is depleted. Crc-mediated regulation impact processes such as biofilm formation, virulence and antibiotic susceptibility.
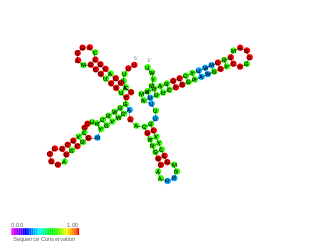
RsmW is a part of the Rsm/Csr family of non-coding RNAs (ncRNAs) discovered in Pseudomonas aeruginosa. It specifically binds to RsmA protein in vitro, restores biofilm production and partially complements the loss of RsmY and RsmZ in rsmY/rsmZ double mutant in regards to their contribution to swarming. Compared to RsmY and RsmZ its production is induced in high temperatures and rsmW is not transcriptionally activated by GacA.
AsponA is a small asRNA transcribed antisense to the penicillin-binding protein 1A gene called ponA. It was identified by RNAseq and the expression was validated by 5' and 3' RACE experiments in Pseudomanas aeruginosa. AsponA expression was up or down regulated under different antibiotic stress. Due to it s location it may be able to prevent the transcription or translation of the opposite gene. Study by Wurtzel et al. and Ferrara et al. also detected its expression.
SrbA is a small regulatory non-coding RNA identified in pathogenic Pseudomonas aeruginosa. It is important for biofilm formation and pathogenicity. Bacterial strain with deleted SrbA had reduced biofilm mass. As the ability to form biofilms can contribute to the ability a pathogen to thrive within the host, the C. elegans hosts infected with the srbA deleted strain displayed significantly lower mortality rate than the wild-type strain. However, the deletion of srbA had no effect on growth or antibiotic resistance in P. aeruginosa.

The ivy-DE RNA motif is a conserved RNA structure that was discovered by bioinformatics. ivy-DE motifs are found in the genus Pseudomonas.















