
In molecular biology, a transcription factor (TF) is a protein that controls the rate of transcription of genetic information from DNA to messenger RNA, by binding to a specific DNA sequence. The function of TFs is to regulate—turn on and off—genes in order to make sure that they are expressed in the right cell at the right time and in the right amount throughout the life of the cell and the organism. Groups of TFs function in a coordinated fashion to direct cell division, cell growth, and cell death throughout life; cell migration and organization during embryonic development; and intermittently in response to signals from outside the cell, such as a hormone. There are up to 1600 TFs in the human genome.

A zinc finger is a small protein structural motif that is characterized by the coordination of one or more zinc ions (Zn2+) in order to stabilize the fold. It was originally coined to describe the finger-like appearance of a hypothesized structure from the African clawed frog (Xenopus laevis) transcription factor IIIA. However, it has been found to encompass a wide variety of differing protein structures in eukaryotic cells. Xenopus laevis TFIIIA was originally demonstrated to contain zinc and require the metal for function in 1983, the first such reported zinc requirement for a gene regulatory protein. It often appears as a metal-binding domain in multi-domain proteins.
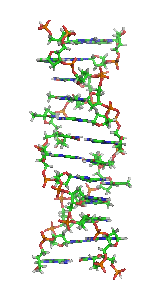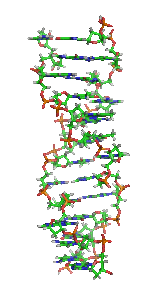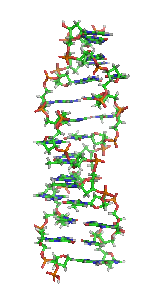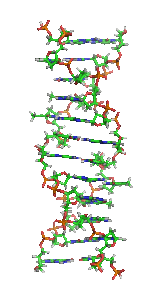
Z-DNA is one of the many possible double helical structures of DNA. It is a left-handed double helical structure in which the helix winds to the left in a zigzag pattern, instead of to the right, like the more common B-DNA form. Z-DNA is thought to be one of three biologically active double-helical structures along with A-DNA and B-DNA.

The glucocorticoid receptor also known as NR3C1 is the receptor to which cortisol and other glucocorticoids bind.

Catabolite activator protein is a trans-acting transcriptional activator that exists as a homodimer in solution. Each subunit of CAP is composed of a ligand-binding domain at the N-terminus and a DNA-binding domain at the C-terminus. Two cAMP molecules bind dimeric CAP with negative cooperativity. Cyclic AMP functions as an allosteric effector by increasing CAP's affinity for DNA. CAP binds a DNA region upstream from the DNA binding site of RNA Polymerase. CAP activates transcription through protein-protein interactions with the α-subunit of RNA Polymerase. This protein-protein interaction is responsible for (i) catalyzing the formation of the RNAP-promoter closed complex; and (ii) isomerization of the RNAP-promoter complex to the open confirmation. CAP's interaction with RNA polymerase causes bending of the DNA near the transcription start site, thus effectively catalyzing the transcription initiation process. CAP's name is derived from its ability to affect transcription of genes involved in many catabolic pathways. For example, when the amount of glucose transported into the cell is low, a cascade of events results in the increase of cytosolic cAMP levels. This increase in cAMP levels is sensed by CAP, which goes on to activate the transcription of many other catabolic genes.

The TATA-binding protein (TBP) is a general transcription factor that binds specifically to a DNA sequence called the TATA box. This DNA sequence is found about 30 base pairs upstream of the transcription start site in some eukaryotic gene promoters.

CREB-binding protein, also known as CREBBP or CBP, is a protein that in humans is encoded by the CREBBP gene. The CREB protein carries out its function by activating transcription, where interaction with transcription factors is managed by one or more CREB domains: the nuclear receptor interaction domain (RID), the KIX domain, the cysteine/histidine regions and the interferon response binding domain (IBiD). The CREB protein domains, KIX, TAZ1 and TAZ2, each bind tightly to a sequence spanning both transactivation domains 9aaTADs of transcription factor p53.

Heterogeneous nuclear ribonucleoprotein A1 is a protein that in humans is encoded by the HNRNPA1 gene. Mutations in hnRNP A1 are causative of amyotrophic lateral sclerosis and the syndrome multisystem proteinopathy.

POU domain, class 2, transcription factor 1 is a protein that in humans is encoded by the POU2F1 gene.

Y box binding protein 1 also known as Y-box transcription factor or nuclease-sensitive element-binding protein 1 is a protein that in humans is encoded by the YBX1 gene.

Transcription factor PU.1 is a protein that in humans is encoded by the SPI1 gene.
RNA-binding protein EWS is a protein that in humans is encoded by the EWSR1 gene on human chromosome 22, specifically 22q12.2. This region of chromosome 22 encodes the N-terminal transactivation domain of the EWS protein and that region may become joined to one of several other chromosomes which encode various transcription factors, see The expression of a chimeric protein with the EWS transactivation domain fused to the DNA binding region of a transcription factor generates a powerful oncogenic protein causing Ewing sarcoma and other members of the Ewing family of tumors. These translocations can occur due to chromoplexy, a burst of complex chromosomal rearrangements seen in cancer cells. The normal EWS gene encodes an RNA binding protein closely related to FUS (gene) and TAF15, all of which have been associated to amyotrophic lateral sclerosis.

Forkhead box protein O4 peptide is a protein that in humans is encoded by the FOXO4 peptide gene. It is located on the long arm of the X chromosome from base pair 71,096,148 to 71,103,533.

RNA-binding protein FUS/TLS, also known as heterogeneous nuclear ribonucleoprotein P2 is a protein that in humans is encoded by the FUS gene.

U1 small nuclear ribonucleoprotein A is a protein that in humans is encoded by the SNRPA gene.

Nuclear transcription factor Y subunit gamma is a protein that in humans is encoded by the NFYC gene.

General transcription factor IIF subunit 1 is a protein that in humans is encoded by the GTF2F1 gene.
General transcription factor IIF subunit 2 is a protein that in humans is encoded by the GTF2F2 gene.
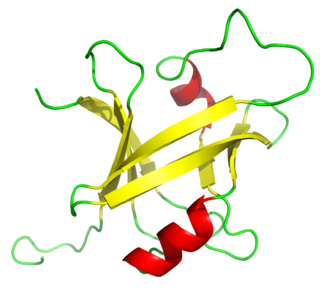
The B3 DNA binding domain (DBD) is a highly conserved domain found exclusively in transcription factors combined with other domains. It consists of 100-120 residues, includes seven beta strands and two alpha helices that form a DNA-binding pseudobarrel protein fold ; it interacts with the major groove of DNA.

General transcription factor IIE subunit 2 (GTF2E2), also known as transcription initiation factor IIE subunit beta (TFIIE-beta), is a protein that in humans is encoded by the GTF2E2 gene.




















