
In neuroscience, long-term potentiation (LTP) is a persistent strengthening of synapses based on recent patterns of activity. These are patterns of synaptic activity that produce a long-lasting increase in signal transmission between two neurons. The opposite of LTP is long-term depression, which produces a long-lasting decrease in synaptic strength.

Pavlovian fear conditioning is a behavioral paradigm in which organisms learn to predict aversive events. It is a form of learning in which an aversive stimulus is associated with a particular neutral context or neutral stimulus, resulting in the expression of fear responses to the originally neutral stimulus or context. This can be done by pairing the neutral stimulus with an aversive stimulus. Eventually, the neutral stimulus alone can elicit the state of fear. In the vocabulary of classical conditioning, the neutral stimulus or context is the "conditional stimulus" (CS), the aversive stimulus is the "unconditional stimulus" (US), and the fear is the "conditional response" (CR).

Brain-derived neurotrophic factor (BDNF), or abrineurin, is a protein that, in humans, is encoded by the BDNF gene. BDNF is a member of the neurotrophin family of growth factors, which are related to the canonical nerve growth factor (NGF), a family which also includes NT-3 and NT-4/NT-5. Neurotrophic factors are found in the brain and the periphery. BDNF was first isolated from a pig brain in 1982 by Yves-Alain Barde and Hans Thoenen.

CREB-TF is a cellular transcription factor. It binds to certain DNA sequences called cAMP response elements (CRE), thereby increasing or decreasing the transcription of the genes. CREB was first described in 1987 as a cAMP-responsive transcription factor regulating the somatostatin gene.

Protein c-Fos is a proto-oncogene that is the human homolog of the retroviral oncogene v-fos. It is encoded in humans by the FOS gene. It was first discovered in rat fibroblasts as the transforming gene of the FBJ MSV. It is a part of a bigger Fos family of transcription factors which includes c-Fos, FosB, Fra-1 and Fra-2. It has been mapped to chromosome region 14q21→q31. c-Fos encodes a 62 kDa protein, which forms heterodimer with c-jun, resulting in the formation of AP-1 complex which binds DNA at AP-1 specific sites at the promoter and enhancer regions of target genes and converts extracellular signals into changes of gene expression. It plays an important role in many cellular functions and has been found to be overexpressed in a variety of cancers.
Ca2+
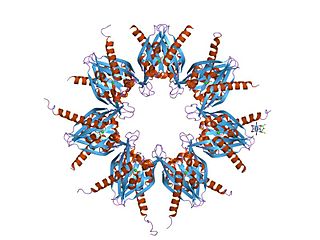
/calmodulin-dependent protein kinase II is a serine/threonine-specific protein kinase that is regulated by the Ca2+
/calmodulin complex. CaMKII is involved in many signaling cascades and is thought to be an important mediator of learning and memory. CaMKII is also necessary for Ca2+
homeostasis and reuptake in cardiomyocytes, chloride transport in epithelia, positive T-cell selection, and CD8 T-cell activation.

Cannabinoid receptor 1 (CB1), is a G protein-coupled cannabinoid receptor that in humans is encoded by the CNR1 gene. The human CB1 receptor is expressed in the peripheral nervous system and central nervous system. It is activated by endogenous cannabinoids called endocannabinoids, a group of retrograde neurotransmitters that include lipids, such as anandamide and 2-arachidonoylglycerol (2-AG); plant phytocannabinoids, such as docosatetraenoylethanolamide found in wild daga, the compound THC which is an active constituent of the psychoactive drug cannabis; and synthetic analogs of THC. CB1 is antagonized by the phytocannabinoid tetrahydrocannabivarin (THCV).
Memory consolidation is a category of processes that stabilize a memory trace after its initial acquisition. A memory trace is a change in the nervous system caused by memorizing something. Consolidation is distinguished into two specific processes. The first, synaptic consolidation, which is thought to correspond to late-phase long-term potentiation, occurs on a small scale in the synaptic connections and neural circuits within the first few hours after learning. The second process is systems consolidation, occurring on a much larger scale in the brain, rendering hippocampus-dependent memories independent of the hippocampus over a period of weeks to years. Recently, a third process has become the focus of research, reconsolidation, in which previously consolidated memories can be made labile again through reactivation of the memory trace.

Activity-regulated cytoskeleton-associated protein is a plasticity protein that in humans is encoded by the ARC gene. The gene is believed to derive from a retrotransposon. The protein is found in the neurons of tetrapods and other animals where it can form virus-like capsids that transport RNA between neurons.
Memory allocation is a process that determines which specific synapses and neurons in a neural network will store a given memory. Although multiple neurons can receive a stimulus, only a subset of the neurons will induce the necessary plasticity for memory encoding. The selection of this subset of neurons is termed neuronal allocation. Similarly, multiple synapses can be activated by a given set of inputs, but specific mechanisms determine which synapses actually go on to encode the memory, and this process is referred to as synaptic allocation. Memory allocation was first discovered in the lateral amygdala by Sheena Josselyn and colleagues in Alcino J. Silva's laboratory.
While the cellular and molecular mechanisms of learning and memory have long been a central focus of neuroscience, it is only in recent years that attention has turned to the epigenetic mechanisms behind the dynamic changes in gene transcription responsible for memory formation and maintenance. Epigenetic gene regulation often involves the physical marking of DNA or associated proteins to cause or allow long-lasting changes in gene activity. Epigenetic mechanisms such as DNA methylation and histone modifications have been shown to play an important role in learning and memory.
Many experiments have been done to find out how the brain interprets stimuli and how animals develop fear responses. The emotion, fear, has been hard-wired into almost every individual, due to its vital role in the survival of the individual. Researchers have found that fear is established unconsciously and that the amygdala is involved with fear conditioning.
The de novo protein synthesis theory of memory formation is a hypothesis about the formation of the physical correlates of memory in the brain. It is widely accepted that the physiological correlates for memories are stored at the synapse between various neurons. The relative strength of various synapses in a network of neurons form the memory trace, or ‘engram,’ though the processes that support this finding are less thoroughly understood. The de novo protein synthesis theory states that the production of proteins is required to initiate and potentially maintain these plastic changes within the brain. It has much support within the neuroscience community, but some critics claim that memories can be made independent of protein synthesis.

Alcino J. Silva is a Portuguese-American neuroscientist who was the recipient of the 2008 Order of Prince Henry and elected as a fellow of the American Association for the Advancement of Science in 2013 for his contributions to the molecular cellular cognition of memory, a field he pioneered with the publication of two articles in Science in 1992.

Kaang Bong-Kiun is a South Korean professor of neuroscience in the Department of Biological Sciences of Seoul National University. He is a fellow of the Korean Academy of Science and Technology and co-director of the IBS Center for Cognition and Sociality with Changjoon Justin Lee.

Memory is commonly referred to as the ability to encode, store, retain and subsequently recall information and past experiences in the human brain. This process involves many proteins, one of which is the Histone-binding protein RbAp48, encoded by the RBBP4 gene in humans.
Epigenetics of depression is the study of how epigenetics contribute to depression.
Alcoholism is a chronic disease characterized by trouble controlling the consumption of alcohol, dependence, and withdrawal upon rapid cessation of drinking. According to ARDI reports approximately 88,000 people had alcohol-related deaths in the United States between the years of 2006 and 2010. Furthermore, chronic alcohol use is consistently the third leading cause of death in the United States. In consequence, research has sought to determine the factors responsible for the development and persistence of alcoholism. From this research, several molecular and epigenetic mechanisms have been discovered.
Christine Denny is an American neuroscientist and associate professor of Clinical Neurobiology in Psychiatry in the Department of Psychiatry at Columbia University Irving Medical Center in New York City. Denny investigates the molecular mechanisms underlying learning and memory. She developed a novel technique to label neurons that encode specific memories. She used this technique to probe what happens to hippocampal memory traces in different disease states.

The TET enzymes are a family of ten-eleven translocation (TET) methylcytosine dioxygenases. They are instrumental in DNA demethylation. 5-Methylcytosine is a methylated form of the DNA base cytosine (C) that often regulates gene transcription and has several other functions in the genome.