Related Research Articles

Microtubules are polymers of tubulin that form part of the cytoskeleton and provide structure and shape to eukaryotic cells. Microtubules can be as long as 50 micrometres, as wide as 23 to 27 nm and have an inner diameter between 11 and 15 nm. They are formed by the polymerization of a dimer of two globular proteins, alpha and beta tubulin into protofilaments that can then associate laterally to form a hollow tube, the microtubule. The most common form of a microtubule consists of 13 protofilaments in the tubular arrangement.

In cell biology, the spindle apparatus is the cytoskeletal structure of eukaryotic cells that forms during cell division to separate sister chromatids between daughter cells. It is referred to as the mitotic spindle during mitosis, a process that produces genetically identical daughter cells, or the meiotic spindle during meiosis, a process that produces gametes with half the number of chromosomes of the parent cell.
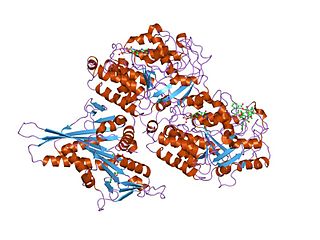
Tubulin in molecular biology can refer either to the tubulin protein superfamily of globular proteins, or one of the member proteins of that superfamily. α- and β-tubulins polymerize into microtubules, a major component of the eukaryotic cytoskeleton. It was discovered and named by Hideo Mōri in 1968. Microtubules function in many essential cellular processes, including mitosis. Tubulin-binding drugs kill cancerous cells by inhibiting microtubule dynamics, which are required for DNA segregation and therefore cell division.
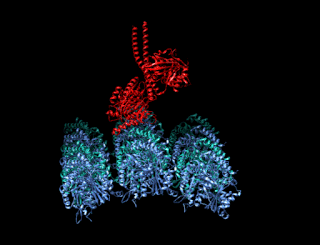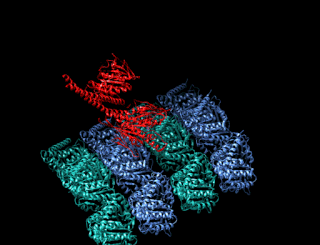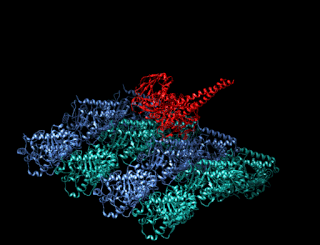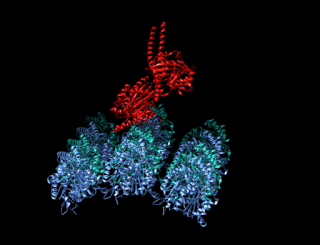
A kinesin is a protein belonging to a class of motor proteins found in eukaryotic cells. Kinesins move along microtubule (MT) filaments and are powered by the hydrolysis of adenosine triphosphate (ATP). The active movement of kinesins supports several cellular functions including mitosis, meiosis and transport of cellular cargo, such as in axonal transport, and intraflagellar transport. Most kinesins walk towards the plus end of a microtubule, which, in most cells, entails transporting cargo such as protein and membrane components from the center of the cell towards the periphery. This form of transport is known as anterograde transport. In contrast, dyneins are motor proteins that move toward the minus end of a microtubule in retrograde transport.

A kinetochore is a disc-shaped protein structure associated with duplicated chromatids in eukaryotic cells where the spindle fibers attach during cell division to pull sister chromatids apart. The kinetochore assembles on the centromere and links the chromosome to microtubule polymers from the mitotic spindle during mitosis and meiosis. The term kinetochore was first used in a footnote in a 1934 Cytology book by Lester W. Sharp and commonly accepted in 1936. Sharp's footnote reads: "The convenient term kinetochore has been suggested to the author by J. A. Moore", likely referring to John Alexander Moore who had joined Columbia University as a freshman in 1932.
In cell biology, microtubule-associated proteins (MAPs) are proteins that interact with the microtubules of the cellular cytoskeleton. MAPs are integral to the stability of the cell and its internal structures and the transport of components within the cell.

Motor proteins are a class of molecular motors that can move along the cytoskeleton of cells. They convert chemical energy into mechanical work by the hydrolysis of ATP. Flagellar rotation, however, is powered by a proton pump.

Stathmin, also known as metablastin and oncoprotein 18 is a protein that in humans is encoded by the STMN1 gene.
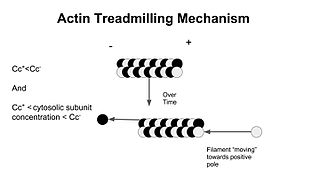
In molecular biology, treadmilling is a phenomenon observed within protein filaments of the cytoskeletons of many cells, especially in actin filaments and microtubules. It occurs when one end of a filament grows in length while the other end shrinks, resulting in a section of filament seemingly "moving" across a stratum or the cytosol. This is due to the constant removal of the protein subunits from these filaments at one end of the filament, while protein subunits are constantly added at the other end. Treadmilling was discovered by Wegner, who defined the thermodynamic and kinetic constraints. Wegner recognized that: “The equilibrium constant (K) for association of a monomer with a polymer is the same at both ends, since the addition of a monomer to each end leads to the same polymer.”; a simple reversible polymer can’t treadmill; ATP hydrolysis is required. GTP is hydrolyzed for microtubule treadmilling.

Aurora kinase B is a protein that functions in the attachment of the mitotic spindle to the centromere and in cytokinesis.
Polo-like kinases (Plks) are regulatory serine/threonine kinases of the cell cycle involved in mitotic entry, mitotic exit, spindle formation, cytokinesis, and meiosis. Only one Plk is found in the genomes of the fly Drosophila melanogaster (Polo), budding yeast (Cdc5) and fission yeast (Plo1). Vertebrates and other animals, however, have many Plk family members including Plk1, Plk2/Snk, Plk3/Prk/FnK, Plk4/Sak and Plk5. Of the vertebrate Plk family members, the mammalian Plk1 has been most extensively studied. During mitosis and cytokinesis, Plks associate with several structures including the centrosome, kinetochores, and the central spindle.

Microtubule-associated protein 4 is a protein that in humans is encoded by the MAP4 gene.
Tubulin alpha-4A chain is a protein that in humans is encoded by the TUBA4A gene.
Tubulin alpha-1A chain is a protein that in humans is encoded by the TUBA1A gene.

Targeting protein for Xklp2 is a protein that in humans is encoded by the TPX2 gene. It is one of the many spindle assembly factors that play a key role in inducing microtubule assembly and growth during M phase.

Kinesin-like protein KIF2C is a protein that in humans is encoded by the KIF2C gene.

Kinesin-like protein KIF2A is a protein that in humans is encoded by the KIF2A gene. In mice, KIF2A is essential for proper neurogenesis and deficiency of KIF2A in mature neurons results in the loss of those neurons.
The XMAP215/Dis1 family is a highly conserved group of microtubule-associated proteins (MAPs) in eukaryotic organisms. These proteins are unique MAPs because they primarily interact with the growing-end (plus-end) of microtubules. This special property classifies this protein family as plus-end tracking proteins (+TIPs).

Neurotubules are microtubules found in neurons in nervous tissues. Along with neurofilaments and microfilaments, they form the cytoskeleton of neurons. Neurotubules are undivided hollow cylinders that are made up of tubulin protein polymers and arrays parallel to the plasma membrane in neurons. Neurotubules have an outer diameter of about 23 nm and an inner diameter, also known as the central core, of about 12 nm. The wall of the neurotubules is about 5 nm in width. There is a non-opaque clear zone surrounding the neurotubule and it is about 40 nm in diameter. Like microtubules, neurotubules are greatly dynamic and the length of them can be adjusted by polymerization and depolymerization of tubulin.
J. Richard McIntosh is a Distinguished Professor Emeritus in Molecular, Cellular, and Developmental Biology at the University of Colorado Boulder. McIntosh first graduated from Harvard with a BA in Physics in 1961, and again with a Ph.D. in Biophysics in 1968. He began his teaching career at Harvard but has spent most of his career at the University of Colorado Boulder. At the University of Colorado Boulder, McIntosh taught biology courses at both the undergraduate and graduate levels. Additionally, he created an undergraduate course in the biology of cancer towards the last several years of his teaching career. McIntosh's research career looks at a variety of things, including different parts of mitosis, microtubules, and motor proteins.
References
- 1 2 3 4 Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P (January 2002). "Mitosis". Molecular Biology of the Cell (4th ed.). New York: Garland Science. ISBN 978-0-7167-3706-3.
- ↑ Lodish H, Berk A, Zipursky SL, Matsudaira P, Baltimore D, Darnell J (January 2000). "Microtubule Structures". Molecular Cell Biology (4th ed.). ISBN 978-0-7167-4366-8.
- ↑ Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P (January 2002). "The Self-Assembly and Dynamic Structure of Cytoskeletal Filaments". Molecular Biology of the Cell (4th ed.). New York: Garland Science. ISBN 978-0-7167-3706-3.
- ↑ Lodish H, Berk A, Zipursky SL, Matsudaira P, Baltimore D, Darnell J (January 2000). "Microtubule Dynamics and Associated Proteins". Molecular Biology of the Cell (4th ed.). New York: Garland Science. ISBN 978-0-7167-3706-3.
- 1 2 3 4 Bowne-Anderson H, Hibbel A, Howard J (December 2015). "Regulation of Microtubule Growth and Catastrophe: Unifying Theory and Experiment". Trends in Cell Biology. 25 (12): 769–779. doi:10.1016/j.tcb.2015.08.009. PMC 4783267 . PMID 26616192.
- ↑ Marklund U, Larsson N, Gradin HM, Brattsand G, Gullberg M (October 1996). "Oncoprotein 18 is a phosphorylation-responsive regulator of microtubule dynamics". The EMBO Journal. 15 (19): 5290–5298. doi:10.1002/j.1460-2075.1996.tb00914.x. PMC 452273 . PMID 8895574.
- 1 2 Brattsand G (August 2000). "Correlation of oncoprotein 18/stathmin expression in human breast cancer with established prognostic factors". British Journal of Cancer. 83 (3): 311–318. doi:10.1054/bjoc.2000.1264. PMC 2374559 . PMID 10917544.
- ↑ Larsson N, Marklund U, Gradin HM, Brattsand G, Gullberg M (September 1997). "Control of microtubule dynamics by oncoprotein 18: dissection of the regulatory role of multisite phosphorylation during mitosis". Molecular and Cellular Biology. 17 (9): 5530–5539. doi:10.1128/MCB.17.9.5530. PMC 232401 . PMID 9271428.
- ↑ Hunter AW, Caplow M, Coy DL, Hancock WO, Diez S, Wordeman L, Howard J (February 2003). "The kinesin-related protein MCAK is a microtubule depolymerase that forms an ATP-hydrolyzing complex at microtubule ends". Molecular Cell. 11 (2): 445–457. doi:10.1016/S1097-2765(03)00049-2. PMC 6468321 . PMID 12620232.
- ↑ Helenius J, Brouhard G, Kalaidzidis Y, Diez S, Howard J (May 2006). "The depolymerizing kinesin MCAK uses lattice diffusion to rapidly target microtubule ends". Nature. 441 (7089): 115–119. Bibcode:2006Natur.441..115H. CiteSeerX 10.1.1.392.1014 . doi:10.1038/nature04736. PMID 16672973. S2CID 4408328.
- ↑ Moriwaki T, Goshima G (November 2016). "Five factors can reconstitute all three phases of microtubule polymerization dynamics". The Journal of Cell Biology. 215 (3): 357–368. doi:10.1083/jcb.201604118. PMC 5100292 . PMID 27799364.