Related Research Articles

Functional magnetic resonance imaging or functional MRI (fMRI) measures brain activity by detecting changes associated with blood flow. This technique relies on the fact that cerebral blood flow and neuronal activation are coupled. When an area of the brain is in use, blood flow to that region also increases.

Neurofeedback is a form of biofeedback that uses electrical potentials in the brain to reinforce desired brain states through operant conditioning. This process is non-invasive and typically collects brain activity data using electroencephalography (EEG). Several neurofeedback protocols exist, with potential additional benefit from use of quantitative electroencephalography (QEEG) or functional magnetic resonance imaging (fMRI) to localize and personalize treatment. Related technologies include functional near-infrared spectroscopy-mediated (fNIRS) neurofeedback, hemoencephalography biofeedback (HEG), and fMRI biofeedback.

A brain–computer interface (BCI), sometimes called a brain–machine interface (BMI) or smartbrain, is a direct communication pathway between the brain's electrical activity and an external device, most commonly a computer or robotic limb. BCIs are often directed at researching, mapping, assisting, augmenting, or repairing human cognitive or sensory-motor functions. They are often conceptualized as a human–machine interface that skips the intermediary component of the physical movement of body parts, although they also raise the possibility of the erasure of the discreteness of brain and machine. Implementations of BCIs range from non-invasive and partially invasive to invasive, based on how close electrodes get to brain tissue.

Discovery science is a scientific methodology which aims to find new patterns, correlations, and form hypotheses through the analysis of large-scale experimental data. The term “discovery science” encompasses various fields of study, including basic, translational, and computational science and research. Discovery-based methodologies are commonly contrasted with traditional scientific practice, the latter involving hypothesis formation before experimental data is closely examined. Discovery science involves the process of inductive reasoning or using observations to make generalisations, and can be applied to a range of science-related fields, e.g., medicine, proteomics, hydrology, psychology, and psychiatry.

Neural oscillations, or brainwaves, are rhythmic or repetitive patterns of neural activity in the central nervous system. Neural tissue can generate oscillatory activity in many ways, driven either by mechanisms within individual neurons or by interactions between neurons. In individual neurons, oscillations can appear either as oscillations in membrane potential or as rhythmic patterns of action potentials, which then produce oscillatory activation of post-synaptic neurons. At the level of neural ensembles, synchronized activity of large numbers of neurons can give rise to macroscopic oscillations, which can be observed in an electroencephalogram. Oscillatory activity in groups of neurons generally arises from feedback connections between the neurons that result in the synchronization of their firing patterns. The interaction between neurons can give rise to oscillations at a different frequency than the firing frequency of individual neurons. A well-known example of macroscopic neural oscillations is alpha activity.
Psychiatric epidemiology is a field which studies the causes (etiology) of mental disorders in society, as well as conceptualization and prevalence of mental illness. It is a subfield of the more general epidemiology. It has roots in sociological studies of the early 20th century. However, while sociological exposures are still widely studied in psychiatric epidemiology, the field has since expanded to the study of a wide area of environmental risk factors, such as major life events, as well as genetic exposures. Increasingly neuroscientific techniques like MRI are used to explore the mechanisms behind how exposures to risk factors may impact psychological problems and explore the neuroanatomical substrate underlying psychiatric disorders.
Brain-reading or thought identification uses the responses of multiple voxels in the brain evoked by stimulus then detected by fMRI in order to decode the original stimulus. Advances in research have made this possible by using human neuroimaging to decode a person's conscious experience based on non-invasive measurements of an individual's brain activity. Brain reading studies differ in the type of decoding employed, the target, and the decoding algorithms employed.
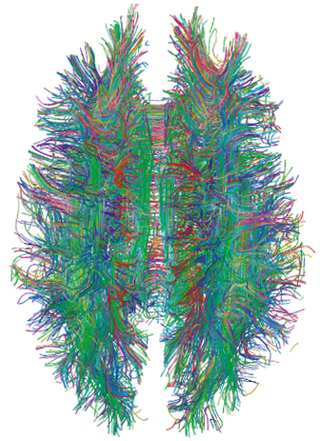
A connectome is a comprehensive map of neural connections in the brain, and may be thought of as its "wiring diagram". An organism's nervous system is made up of neurons which communicate through synapses. A connectome is constructed by tracing the neuron in a nervous system and mapping where neurons are connected through synapses.

National Brain Research Centre is a research institute in Manesar, Gurgaon, India. It is an autonomous institute under the Department of Biotechnology, Ministry of Science and Technology, Government of India. The institute is dedicated to research in neuroscience and brain functions in health and diseases using multidisciplinary approaches. This is the first autonomous institute by DBT to be awarded by the Ministry of Education, Government of India, formerly known as the Ministry of Human Resource Development, in May 2002. NBRC was dedicated to the nation by the Honorable President of India Dr. A.P.J. Abdul Kalam in December 2003. The founder chairman of NBRC Society is Prof. Prakash Narain Tandon, whereas the founder director Prof. Vijayalakshmi Ravindranath was followed by Prof. Subrata Sinha and Prof. Neeraj Jain. The current director of NBRC is Prof. Krishanu Ray.
Connectomics is the production and study of connectomes: comprehensive maps of connections within an organism's nervous system. More generally, it can be thought of as the study of neuronal wiring diagrams with a focus on how structural connectivity, individual synapses, cellular morphology, and cellular ultrastructure contribute to the make up of a network. The nervous system is a network made of billions of connections and these connections are responsible for our thoughts, emotions, actions, memories, function and dysfunction. Therefore, the study of connectomics aims to advance our understanding of mental health and cognition by understanding how cells in the nervous system are connected and communicate. Because these structures are extremely complex, methods within this field use a high-throughput application of functional and structural neural imaging, most commonly magnetic resonance imaging (MRI), electron microscopy, and histological techniques in order to increase the speed, efficiency, and resolution of these nervous system maps. To date, tens of large scale datasets have been collected spanning the nervous system including the various areas of cortex, cerebellum, the retina, the peripheral nervous system and neuromuscular junctions.
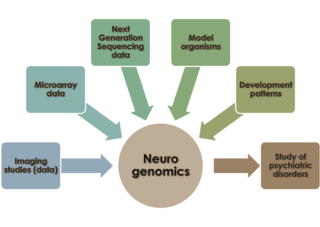
Neurogenomics is the study of how the genome of an organism influences the development and function of its nervous system. This field intends to unite functional genomics and neurobiology in order to understand the nervous system as a whole from a genomic perspective.
Imagined speech is thinking in the form of sound – "hearing" one's own voice silently to oneself, without the intentional movement of any extremities such as the lips, tongue, or hands. Logically, imagined speech has been possible since the emergence of language, however, the phenomenon is most associated with its investigation through signal processing and detection within electroencephalograph (EEG) data as well as data obtained using alternative non-invasive, brain–computer interface (BCI) devices.
Medical image computing (MIC) is an interdisciplinary field at the intersection of computer science, information engineering, electrical engineering, physics, mathematics and medicine. This field develops computational and mathematical methods for solving problems pertaining to medical images and their use for biomedical research and clinical care.

Resting state fMRI is a method of functional magnetic resonance imaging (fMRI) that is used in brain mapping to evaluate regional interactions that occur in a resting or task-negative state, when an explicit task is not being performed. A number of resting-state brain networks have been identified, one of which is the default mode network. These brain networks are observed through changes in blood flow in the brain which creates what is referred to as a blood-oxygen-level dependent (BOLD) signal that can be measured using fMRI.
Dynamic functional connectivity (DFC) refers to the observed phenomenon that functional connectivity changes over a short time. Dynamic functional connectivity is a recent expansion on traditional functional connectivity analysis which typically assumes that functional networks are static in time. DFC is related to a variety of different neurological disorders, and has been suggested to be a more accurate representation of functional brain networks. The primary tool for analyzing DFC is fMRI, but DFC has also been observed with several other mediums. DFC is a recent development within the field of functional neuroimaging whose discovery was motivated by the observation of temporal variability in the rising field of steady state connectivity research.

Russell "Russ" Alan Poldrack is an American psychologist and neuroscientist. He is a professor of psychology at Stanford University, associate director of Stanford Data Science, member of the Stanford Neuroscience Institute and director of the Stanford Center for Reproducible Neuroscience and the SDS Center for Open and Reproducible Science.
In the field of medicine, radiomics is a method that extracts a large number of features from medical images using data-characterisation algorithms. These features, termed radiomic features, have the potential to uncover tumoral patterns and characteristics that fail to be appreciated by the naked eye. The hypothesis of radiomics is that the distinctive imaging features between disease forms may be useful for predicting prognosis and therapeutic response for various cancer types, thus providing valuable information for personalized therapy. Radiomics emerged from the medical fields of radiology and oncology and is the most advanced in applications within these fields. However, the technique can be applied to any medical study where a pathological process can be imaged.
James Van Loan Haxby is an American neuroscientist. He currently is a professor in the Department of Psychological and Brain Sciences at Dartmouth College and was the Director for the Dartmouth Center for Cognitive Neuroscience from 2008 to 2021. He is best known for his work on face perception and applications of machine learning in functional neuroimaging.
Dynamic causal modeling (DCM) is a framework for specifying models, fitting them to data and comparing their evidence using Bayesian model comparison. It uses nonlinear state-space models in continuous time, specified using stochastic or ordinary differential equations. DCM was initially developed for testing hypotheses about neural dynamics. In this setting, differential equations describe the interaction of neural populations, which directly or indirectly give rise to functional neuroimaging data e.g., functional magnetic resonance imaging (fMRI), magnetoencephalography (MEG) or electroencephalography (EEG). Parameters in these models quantify the directed influences or effective connectivity among neuronal populations, which are estimated from the data using Bayesian statistical methods.

John-Dylan Haynes is a British-German brain researcher.
References
- Kriegeskorte, N.; Simmons, W. K.; Bellgowan, P. S. F.; Baker, C. I. (2009). "Circular analysis in systems neuroscience: The dangers of double dipping". Nature Neuroscience. 12 (5): 535–540. doi:10.1038/nn.2303. PMC 2841687 . PMID 19396166.
- Kriegeskorte, N.; Lindquist, M. A.; Nichols, T. E.; Poldrack, R. A.; Vul, E. (2010). "Everything you never wanted to know about circular analysis, but were afraid to ask". Journal of Cerebral Blood Flow & Metabolism. 30 (9): 1551–1557. doi:10.1038/jcbfm.2010.86. PMC 2949251 . PMID 20571517.
- Tolstrup, N.; Rouzé, P.; Brunak, S. (1997). "A branch point consensus from Arabidopsis found by non-circular analysis allows for better prediction of acceptor sites". Nucleic Acids Research. 25 (15): 3159–3163. doi:10.1093/nar/25.15.3159. PMC 146848 . PMID 9224618.
- Olivetti, E.; Mognon, A.; Greiner, S.; Avesani, P. (2010). "Brain Decoding: Biases in Error Estimation". 2010 First Workshop on Brain Decoding: Pattern Recognition Challenges in Neuroimaging. pp. 40–43. doi:10.1109/WBD.2010.9. ISBN 978-1-4244-8486-7. S2CID 13024947.