
Cellulase is any of several enzymes produced chiefly by fungi, bacteria, and protozoans that catalyze cellulolysis, the decomposition of cellulose and of some related polysaccharides. The name is also used for any naturally occurring mixture or complex of various such enzymes, that act serially or synergistically to decompose cellulosic material.

Separase, also known as separin, is a cysteine protease responsible for triggering anaphase by hydrolysing cohesin, which is the protein responsible for binding sister chromatids during the early stage of anaphase. In humans, separin is encoded by the ESPL1 gene.
SMC complexes represent a large family of ATPases that participate in many aspects of higher-order chromosome organization and dynamics. SMC stands for Structural Maintenance of Chromosomes.
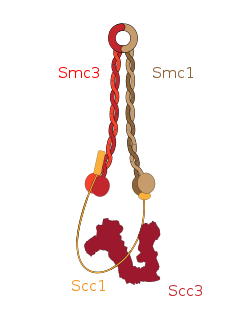
Cohesin is a protein complex that mediates sister chromatid cohesion, homologous recombination and DNA looping. Cohesin is formed of SMC3, SMC1, SCC1 and SCC3. Cohesin holds sister chromatids together after DNA replication until anaphase when removal of cohesin leads to separation of sister chromatids. The complex forms a ring-like structure and it is believed that sister chromatids are held together by entrapment inside the cohesin ring. Cohesin is a member of the SMC family of protein complexes which includes Condensin, MukBEF and SMC-ScpAB.
Cellulosomes are multi-enzyme extracellular complexes. Cellulosomes are associated with the cell surface and mediate cell attachment to insoluble substrates and degrade them to soluble products which are then absorbed. Cellulosome complexes are intricate, multi-enzyme machines, produced by many cellulolytic microorganisms. They are produced by microorganisms for efficient degradation of plant cell wall polysaccharides, notably cellulose, the most abundant organic polymer on Earth. The multiple subunits of cellulosomes are composed of numerous functional domains that interact with each other and with the cellulosic substrate. One of these subunits, a large glycoprotein "scaffoldin", is a distinctive class of non-catalytic scaffolding polypeptides. The scaffoldin subunit selectively integrates the various cellulases and xylanase subunits into the cohesive complex, by combining its cohesin domains with a typical dockerin domain present on each of the subunit enzymes. The scaffoldin of some cellulosomes, an example being that of Clostridium thermocellum, contains a carbohydrate-binding module that adheres cellulose to the cellulosomal complex.

The EF hand is a helix–loop–helix structural domain or motif found in a large family of calcium-binding proteins.
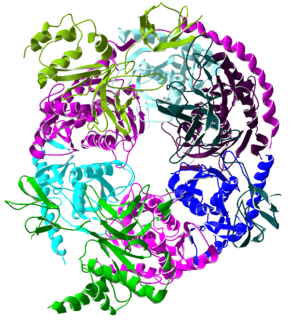
The exosome complex is a multi-protein intracellular complex capable of degrading various types of RNA molecules. Exosome complexes are found in both eukaryotic cells and archaea, while in bacteria a simpler complex called the degradosome carries out similar functions.
Clostridium thermocellum is an anaerobic, thermophilic bacterium. C. thermocellum has garnered research interest due to its cellulolytic and ethanologenic abilities, being capable of directly converting a cellulosic substrate into ethanol by consolidated bioprocessing. This makes it useful in converting biomass into a usable energy source. The degradation of the cellulose is carried out in the bacterium by a large extracellular cellulase system called a cellulosome, which contains nearly 20 catalytic subunits. The cellulase system of the bacterium significantly differs from fungal cellulases due to its high activity on crystalline cellulose, being able to completely solubilize crystalline sources of cellulose, such as cotton. However, there are some shortfalls in applying the organism to practical applications due to it having low ethanol yield, at least partially due to branched fermentation pathways that produce acetate, formate, and lactate along with ethanol. There is also evidence of inhibition due to the presence of hydrogen and due to agitation. Some recent research has been directed to optimizing the ethanol-producing metabolic pathway in hopes of creating more efficient biomass conversion.

Nipped-B-like protein (NIPBL), also known as SCC2 or delangin is a protein that in humans is encoded by the NIPBL gene. NIPBL is required for the association of cohesin with DNA and is the major subunit of the cohesin loading complex. Heterozygous mutations in NIPBL account for an estimated 60% of case of Cornelia de Lange Syndrome.

Structural maintenance of chromosomes protein 1A (SMC1A) is a protein that in humans is encoded by the SMC1A gene. SMC1A is a subunit of the cohesin complex which mediates sister chromatid cohesion, homologous recombination and DNA looping. In somatic cells, cohesin is formed of SMC1A, SMC3, RAD21 and either SA1 or SA2 whereas in meiosis, cohesin is formed of SMC3, SMC1B, REC8 and SA3.

RNase PH is a tRNA nucleotidyltransferase, present in archaea and bacteria, that is involved in tRNA processing. Contrary to hydrolytic enzymes, it is a phosphorolytic enzyme, meaning that it uses inorganic phosphate as a reactant to cleave nucleotide-nucleotide bonds, releasing diphosphate nucleotides. The active structure of the proteins is a homohexameric complex, consisting of three ribonuclease (RNase) PH dimers. RNase PH has homologues in many other organisms, which are referred to as RNase PH-like proteins. The part of another larger protein with a domain that is very similar to RNase PH is called an RNase PH domain (RPD).

Structural maintenance of chromosomes protein 3 (SMC3) is a protein that in humans is encoded by the SMC3 gene. SMC3 is a subunit of the Cohesin complex which mediates sister chromatid cohesion, homologous recombination and DNA looping. Cohesin is formed of SMC3, SMC1, RAD21 and either SA1 or SA2. In humans, SMC3 is present in all cohesin complexes whereas there are multiple paralogs for the other subunits.

Wings apart-like protein homolog (WAPL) is a protein that in humans is encoded by the WAPAL gene. WAPL is a key regulator of the Cohesin complex which mediates sister chromatid cohesion, homologous recombination and DNA looping. Cohesin is formed of SMC3, SMC1, RAD21 and either SA1 or SA2. Cohesin has a ring-like arrangement and it is thought that it associates with the chromosome by entrapping it whether as a loop of DNA, a single strand or a pair of sister chromosomes. WAPL forms a complex with PDS5A or PDS5B and releases cohesin from DNA by opening the interface between SMC3 and RAD21.

Dockerin is a protein domain found in the cellulosome cellular structure of anaerobic bacteria. It is found on many endoglucanase enzymes. The dockerin's binding partner is the cohesin domain, located on the scaffoldin protein. This interaction between the dockerin domains of the enzyme constituents of the cellulosome and the cohesin domains of the scaffoldin protein is essential to the construction of the cellulosome complex. The Dockerin domain has two in-tandem repeats of a non-EF hand calcium binding motif. Each motif is characterized by a loop-helix structure. The three-dimensional structure of dockerin has been determined in solution, as well as in complex with Cohesin.

A HEAT repeat is a protein tandem repeat structural motif composed of two alpha helices linked by a short loop. HEAT repeats can form alpha solenoids, a type of solenoid protein domain found in a number of cytoplasmic proteins. The name "HEAT" is an acronym for four proteins in which this repeat structure is found: Huntingtin, elongation factor 3 (EF3), protein phosphatase 2A (PP2A), and the yeast kinase TOR1. HEAT repeats form extended superhelical structures which are often involved in intracellular transport; they are structurally related to armadillo repeats. The nuclear transport protein importin beta contains 19 HEAT repeats.
Sister chromatid cohesion refers to the process by which sister chromatids are paired and held together during certain phases of the cell cycle. Establishment of sister chromatid cohesion is the process by which chromatin-associated cohesin protein becomes competent to physically bind together the sister chromatids. In general, cohesion is established during S phase as DNA is replicated, and is lost when chromosomes segregate during mitosis and meiosis. Some studies have suggested that cohesion aids in aligning the kinetochores during mitosis by forcing the kinetochores to face opposite cell poles.

In molecular biology, a carbohydrate-binding module (CBM) is a protein domain found in carbohydrate-active enzymes. The majority of these domains have carbohydrate-binding activity. Some of these domains are found on cellulosomal scaffoldin proteins. CBMs were previously known as cellulose-binding domains. CBMs are classified into numerous families, based on amino acid sequence similarity. There are currently 64 families of CBM in the CAZy database.
Cellulose 1,4-beta-cellobiosidase is an enzyme with systematic name 4-beta-D-glucan cellobiohydrolase . This enzyme catalyses the following chemical reaction
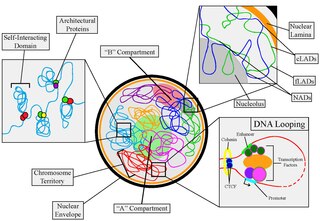
Nuclear organization refers to the spatial distribution of chromatin within a cell nucleus. There are many different levels and scales of nuclear organisation. Chromatin is a higher order structure of DNA.

Edward A. Bayer is an American-Israeli scientist.















