
Cellulase is any of several enzymes produced chiefly by fungi, bacteria, and protozoans that catalyze cellulolysis, the decomposition of cellulose and of some related polysaccharides. The name is also used for any naturally occurring mixture or complex of various such enzymes, that act serially or synergistically to decompose cellulosic material.
In biology and biochemistry, protease inhibitors, or antiproteases, are molecules that inhibit the function of proteases. Many naturally occurring protease inhibitors are proteins.

Nidogen-1 (NID-1), formerly known as entactin, is a protein that in humans is encoded by the NID1 gene. Both nidogen-1 and nidogen-2 are essential components of the basement membrane alongside other components such as type IV collagen, proteoglycans, laminin and fibronectin.

Catabolite activator protein is a trans-acting transcriptional activator that exists as a homodimer in solution. Each subunit of CAP is composed of a ligand-binding domain at the N-terminus and a DNA-binding domain at the C-terminus. Two cAMP molecules bind dimeric CAP with negative cooperativity. Cyclic AMP functions as an allosteric effector by increasing CAP's affinity for DNA. CAP binds a DNA region upstream from the DNA binding site of RNA Polymerase. CAP activates transcription through protein-protein interactions with the α-subunit of RNA Polymerase. This protein-protein interaction is responsible for (i) catalyzing the formation of the RNAP-promoter closed complex; and (ii) isomerization of the RNAP-promoter complex to the open confirmation. CAP's interaction with RNA polymerase causes bending of the DNA near the transcription start site, thus effectively catalyzing the transcription initiation process. CAP's name is derived from its ability to affect transcription of genes involved in many catabolic pathways. For example, when the amount of glucose transported into the cell is low, a cascade of events results in the increase of cytosolic cAMP levels. This increase in cAMP levels is sensed by CAP, which goes on to activate the transcription of many other catabolic genes.
Cellulosomes are multi-enzyme complexes. Cellulosomes are associated with the cell surface and mediate cell attachment to insoluble substrates and degrade them to soluble products which are then absorbed. Cellulosome complexes are intricate, multi-enzyme machines, produced by many cellulolytic microorganisms. They are produced by microorganisms for efficient degradation of plant cell wall polysaccharides, notably cellulose, the most abundant organic polymer on Earth. The multiple subunits of cellulosomes are composed of numerous functional domains which interact with each other and with the cellulosic substrate. One of these subunits, a large glycoprotein "scaffoldin", is a distinctive class of non-catalytic scaffolding polypeptides. The scaffoldin subunit selectively integrates the various cellulases and xylanase subunits into the cohesive complex, by combining its cohesin domains with a typical dockerin domain present on each of the subunit enzymes. The scaffoldin of some cellulosomes, an example being that of Clostridium thermocellum, contains a carbohydrate-binding module that adheres cellulose to the cellulosomal complex.

The EF hand is a helix-loop-helix structural domain or motif found in a large family of calcium-binding proteins.
Acidophiles or acidophilic organisms are those that thrive under highly acidic conditions. These organisms can be found in different branches of the tree of life, including Archaea, Bacteria, and Eukarya.

Transcriptional repressor CTCF also known as 11-zinc finger protein or CCCTC-binding factor is a transcription factor that in humans is encoded by the CTCF gene. CTCF is involved in many cellular processes, including transcriptional regulation, insulator activity, V(D)J recombination and regulation of chromatin architecture.
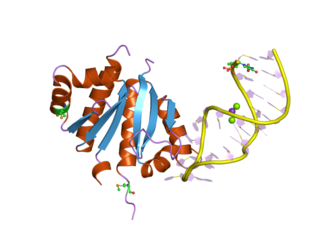
The K Homology (KH) domain is a protein domain that was first identified in the human heterogeneous nuclear ribonucleoprotein (hnRNP) K. An evolutionarily conserved sequence of around 70 amino acids, the KH domain is present in a wide variety of nucleic acid-binding proteins. The KH domain binds RNA, and can function in RNA recognition. It is found in multiple copies in several proteins, where they can function cooperatively or independently. For example, in the AU-rich element RNA-binding protein KSRP, which has 4 KH domains, KH domains 3 and 4 behave as independent binding modules to interact with different regions of the AU-rich RNA targets. The solution structure of the first KH domain of FMR1 and of the C-terminal KH domain of hnRNP K determined by nuclear magnetic resonance (NMR) revealed a beta-alpha-alpha-beta-beta-alpha structure. Autoantibodies to NOVA1, a KH domain protein, cause paraneoplastic opsoclonus ataxia. The KH domain is found at the N-terminus of the ribosomal protein S3. This domain is unusual in that it has a different fold compared to the normal KH domain.

Heterogeneous nuclear ribonucleoprotein K is a protein that in humans is encoded by the HNRNPK gene.

FBLN1 is the gene encoding fibulin-1, an extracellular matrix and plasma protein.

Filamin A, alpha (FLNA) is a protein that in humans is encoded by the FLNA gene.

Poly(rC)-binding protein 2 is a protein that in humans is encoded by the PCBP2 gene.
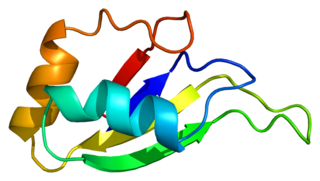
Heterogeneous nuclear ribonucleoprotein D0 (HNRNPD) also known as AU-rich element RNA-binding protein 1 (AUF1) is a protein that in humans is encoded by the HNRNPD gene. Alternative splicing of this gene results in four transcript variants.
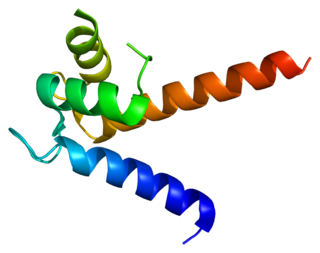
S100 calcium-binding protein P (S100P) is a protein that in humans is encoded by the S100P gene.

Voltage-dependent L-type calcium channel subunit beta-4 is a protein that in humans is encoded by the CACNB4 gene.

In molecular biology, a carbohydrate-binding module (CBM) is a protein domain found in carbohydrate-active enzymes. The majority of these domains have carbohydrate-binding activity. Some of these domains are found on cellulosomal scaffoldin proteins. CBMs were previously known as cellulose-binding domains. CBMs are classified into numerous families, based on amino acid sequence similarity. There are currently 64 families of CBM in the CAZy database.

In molecular biology, the cohesin domain is a protein domain. It interacts with a complementary domain, termed the dockerin domain. The cohesin-dockerin interaction is the crucial interaction for complex formation in the cellulosome.
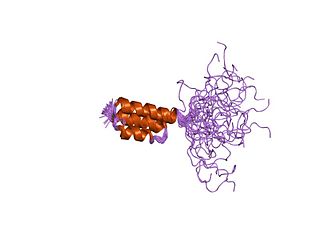
In molecular biology, the GA module, or protein G-related albumin-binding module, is a protein domain which occurs on the surface of numerous Gram-positive bacterial pathogens. Protein G of group C and G Streptococci interacts with the constant region of IgG and with human serum albumin. The GA module is composed of a left-handed three-helix bundle and is found in a range of bacterial cell surface proteins. GA modules may promote bacterial growth and virulence in mammalian hosts by scavenging albumin-bound nutrients and camouflaging the bacteria. Variations in sequence give rise to differences in structure and function between GA modules in different proteins, which could alter pathogenesis and host specificity due to their varied affinities for different species of albumin. Proteins containing a GA module include PAB from Peptostreptococcus magnus.

Edward A. Bayer is an American-Israeli scientist.

















