
The alpha helix (α-helix) is a common motif in the secondary structure of proteins and is a right hand-helix conformation in which every backbone N−H group hydrogen bonds to the backbone C=O group of the amino acid located three or four residues earlier along the protein sequence.
Enterobacteria phage λ is a bacterial virus, or bacteriophage, that infects the bacterial species Escherichia coli. It was discovered by Esther Lederberg in 1950. The wild type of this virus has a temperate life cycle that allows it to either reside within the genome of its host through lysogeny or enter into a lytic phase, during which it kills and lyses the cell to produce offspring. Lambda strains, mutated at specific sites, are unable to lysogenize cells; instead, they grow and enter the lytic cycle after superinfecting an already lysogenized cell.

Protein secondary structure is the three dimensional form of local segments of proteins. The two most common secondary structural elements are alpha helices and beta sheets, though beta turns and omega loops occur as well. Secondary structure elements typically spontaneously form as an intermediate before the protein folds into its three dimensional tertiary structure.

A homeobox is a DNA sequence, around 180 base pairs long, found within genes that are involved in the regulation of patterns of anatomical development (morphogenesis) in animals, fungi, plants, and numerous single cell eukaryotes. Homeobox genes encode homeodomain protein products that are transcription factors sharing a characteristic protein fold structure that binds DNA to regulate expression of target genes. Homeodomain proteins regulate gene expression and cell differentiation during early embryonic development, thus mutations in homeobox genes can cause developmental disorders.
In a chain-like biological molecule, such as a protein or nucleic acid, a structural motif is a supersecondary structure, which also appears in a variety of other molecules. Motifs do not allow us to predict the biological functions: they are found in proteins and enzymes with dissimilar functions.
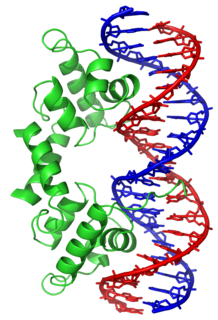
In proteins, the helix-turn-helix (HTH) is a major structural motif capable of binding DNA. Each monomer incorporates two α helices, joined by a short strand of amino acids, that bind to the major groove of DNA. The HTH motif occurs in many proteins that regulate gene expression. It should not be confused with the helix-loop-helix motif.
A supersecondary structure is a compact three-dimensional protein structure of several adjacent elements of a secondary structure that is smaller than a protein domain or a subunit. Supersecondary structures can act as nucleations in the process of protein folding.
A DNA-binding domain (DBD) is an independently folded protein domain that contains at least one structural motif that recognizes double- or single-stranded DNA. A DBD can recognize a specific DNA sequence or have a general affinity to DNA. Some DNA-binding domains may also include nucleic acids in their folded structure.
Tet Repressor proteins are proteins playing an important role in conferring antibiotic resistance to large categories of bacterial species.

A DNA clamp, also known as a sliding clamp or β-clamp, is a protein complex that serves as a processivity-promoting factor in DNA replication. As a critical component of the DNA polymerase III holoenzyme, the clamp protein binds DNA polymerase and prevents this enzyme from dissociating from the template DNA strand. The clamp-polymerase protein–protein interactions are stronger and more specific than the direct interactions between the polymerase and the template DNA strand; because one of the rate-limiting steps in the DNA synthesis reaction is the association of the polymerase with the DNA template, the presence of the sliding clamp dramatically increases the number of nucleotides that the polymerase can add to the growing strand per association event. The presence of the DNA clamp can increase the rate of DNA synthesis up to 1,000-fold compared with a nonprocessive polymerase.
POU is a family of proteins that have well-conserved homeodomains.

Mark Ptashne is a molecular biologist. He is the Ludwig Chair of Molecular Biology at Memorial Sloan–Kettering Cancer Center in New York City.

Brian W. Matthews is a biochemist and biophysicist educated at the University of Adelaide, contributor to x-ray crystallographic methodology at the University of Cambridge, and since 1970 at the University of Oregon as Professor of Physics and HHMI investigator in the Institute of Molecular Biology.

In molecular biology, excisionase is a bacteriophage protein encoded by the Xis gene. It is involved in excisive recombination by regulating the assembly of the excisive intasome and by inhibiting viral integration. It adopts an unusual winged-helix structure in which two alpha helices are packed against two extended strands. Also present in the structure is a two-stranded anti-parallel beta-sheet, whose strands are connected by a four-residue wing. During interaction with DNA, helix alpha2 is thought to insert into the major groove, while the wing contacts the adjacent minor groove or phosphodiester backbone. The C-terminal region of excisionase is involved in interaction with phage-encoded integrase (Int), and a putative C-terminal alpha helix may fold upon interaction with Int and/or DNA.
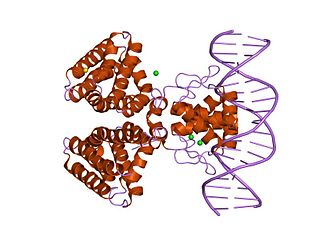
In molecular biology, the fatty acid metabolism regulator protein FadR, is a bacterial transcription factor.

In molecular biology, the iron dependent repressors are a family of bacterial and archaeal transcriptional repressors.

In molecular biology, the LuxR-type DNA-binding HTH domain is a DNA-binding, helix-turn-helix (HTH) domain of about 65 amino acids. It is present in transcription regulators of the LuxR/FixJ family of response regulators. The domain is named after Vibrio fischeri luxR, a transcriptional activator for quorum-sensing control of luminescence. LuxR-type HTH domain proteins occur in a variety of organisms. The DNA-binding HTH domain is usually located in the C-terminal region of the protein; the N-terminal region often containing an autoinducer-binding domain or a response regulatory domain. Most luxR-type regulators act as transcription activators, but some can be repressors or have a dual role for different sites. LuxR-type HTH regulators control a wide variety of activities in various biological processes.

In molecular biology, the GntR-like bacterial transcription factors are a family of transcription factors.
The tailspike protein (P22TSP) of Enterobacteria phage P22 mediates the recognition and adhesion between the bacteriophage and the surface of Salmonella enterica cells. It is anchored within the viral coat and recognizes the O-antigen portion of the lipopolysaccharide (LPS) on the outer-membrane of Gram-negative bacteria. It possesses endoglycanase activity, serving to shorten the length of the O-antigen during infection.

cII or transcriptional activator II is a DNA-binding protein and important transcription factor in the life cycle of lambda phage. It is encoded in the lambda phage genome by the 291 base pair cII gene. cII plays a key role in determining whether the bacteriophage will incorporate its genome into its host and lie dormant (lysogeny), or replicate and kill the host (lysis).
















