Related Research Articles
Chloroform, or trichloromethane, is an organic compound with formula CHCl3. It is a colorless, strong-smelling, dense liquid that is produced on a large scale as a precursor to PTFE. It is also a precursor to various refrigerants. It is one of the four chloromethanes and a trihalomethane. It is a powerful anesthetic, euphoriant, anxiolytic and sedative when inhaled or ingested.
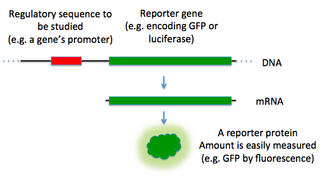
In molecular biology, a reporter gene is a gene that researchers attach to a regulatory sequence of another gene of interest in bacteria, cell culture, animals or plants. Such genes are called reporters because the characteristics they confer on organisms expressing them are easily identified and measured, or because they are selectable markers. Reporter genes are often used as an indication of whether a certain gene has been taken up by or expressed in the cell or organism population.
Reductive dechlorination is Chemical reaction of chlorinated organic compounds with reductants. The reaction breaks C-Cl bonds, and releases chloride ions. Many modalities have been implemented, depending on the application. Reductive dechlorination is often applied to remediation of chlorinated pesticides or dry cleaning solvents. It is also used occasionally in the synthesis of organic compounds, e.g. as pharmaceuticals.
A hydrogenase is an enzyme that catalyses the reversible oxidation of molecular hydrogen (H2), as shown below:
Halorespiration or dehalorespiration or organohalide respiration is the use of halogenated compounds as terminal electron acceptors in anaerobic respiration. Halorespiration can play a part in microbial biodegradation. The most common substrates are chlorinated aliphatics, chlorinated phenols and chloroform. Dehalorespiring bacteria are highly diverse. This trait is found in some proteobacteria, chloroflexi, low G+C gram positive Clostridia. and ultramicrobacteria.
Dehalococcoides is a genus of bacteria within class Dehalococcoidia that obtain energy via the oxidation of hydrogen and subsequent reductive dehalogenation of halogenated organic compounds in a mode of anaerobic respiration called organohalide respiration. They are well known for their great potential to remediate halogenated ethenes and aromatics. They are the only bacteria known to transform highly chlorinated dioxins, PCBs. In addition, they are the only known bacteria to transform tetrachloroethene to ethene.

Cupriavidus necator is a Gram-negative soil bacterium of the class Betaproteobacteria.
Microbial biodegradation is the use of bioremediation and biotransformation methods to harness the naturally occurring ability of microbial xenobiotic metabolism to degrade, transform or accumulate environmental pollutants, including hydrocarbons, polychlorinated biphenyls (PCBs), polyaromatic hydrocarbons (PAHs), heterocyclic compounds, pharmaceutical substances, radionuclides and metals.
In enzymology, a tetrachloroethene reductive dehalogenase is an enzyme that catalyzes the chemical reaction. This is a member of reductive dehalogenase enzyme family.
In enzymology, a (S)-2-haloacid dehalogenase (EC 3.8.1.2) is an enzyme that catalyzes the chemical reaction

Haloacid dehalogenase-like hydrolase domain-containing protein 2 is an enzyme that in humans is encoded by the HDHD2 gene.

γ -L-Glutamyl-L-cysteine, also known as γ-glutamylcysteine (GGC), is a dipeptide found in animals, plants, fungi, some bacteria, and archaea. It has a relatively unusual γ-bond between the constituent amino acids, L-glutamic acid and L-cysteine and is a key intermediate in the gamma (γ) -glutamyl cycle first described by Meister in the 1970s. It is the most immediate precursor to the antioxidant glutathione.
Dehalobacter is a genus in the phylum Firmicutes (Bacteria).

The haloacid dehydrogenase superfamily is a superfamily of enzymes that include phosphatases, phosphonatases, P-type ATPases, beta-phosphoglucomutases, phosphomannomutases, and dehalogenases, and are involved in a variety of cellular processes ranging from amino acid biosynthesis to detoxification.
[NiFe] hydrogenase is a type of hydrogenase, which is an oxidative enzyme that reversibly convert molecular hydrogen in prokaryotes including Bacteria and Archaea. The catalytic site on the enzyme provides simple hydrogen-metabolizing microorganisms a redox mechanism by which to store and utilize energy via the reaction shown in Figure 1. This is particularly essential for the anaerobic, sulfate-reducing bacteria of the genus Desulfovibrio as well as pathogenic organisms Escherichia coli and Helicobacter pylori. The mechanisms, maturation, and function of [NiFe] hydrogenases are actively being researched for applications to the hydrogen economy and as potential antibiotic targets.
Dehalobacter restrictus is a species of bacteria in the phylum Firmicutes. It is strictly anaerobic and reductively dechlorinates tetra- and trichloroethene. It does not form spores; it is a small, gram-positive rod with one lateral flagellum. PER-K23 is its type strain.
Desulfitobacterium dehalogenans is a species of bacteria. They are facultative organohalide respiring bacteria capable of reductively dechlorinating chlorophenolic compounds and tetrachloroethene. They are anaerobic, motile, Gram-positive and rod-shaped bacteria capable of utilizing a wide range of electron donors and acceptors. The type strain JW/IU-DCT, DSM 9161, NCBi taxonomy ID 756499.
Reductive dehaholagenses (EC 1.97.1.8) are a group of enzymes utilized in organohalide respiring bacteria. These enzymes are mostly attached to the periplasmic side of the cytoplasmic membrane and play a central role in energy-conserving respiratory process for organohalide respiring bacteria by reducing organohalides. During such reductive dehalogenation reaction, organohalides are used as terminal electron acceptors. They catalyze the following general reactions:
Jagnyeswar Ratha is a professor at the School of Life Sciences, Sambalpur University, Burla. He teaches biochemistry, cell biology, immunology and animal biotechnology.
Sam Hay is a chemist from New Zealand and a Reader in the Department of Chemistry at The University of Manchester. His research in general is based on computational chemistry and theoretical chemistry, specifically on the areas of In silico Enzymology, quantum mechanics roles in biological processes, kinetic modelling of complex reactions and high pressure spectroscopy.
References
- ↑ Jugder, Bat-Erdene; Ertan, Haluk; Lee, Matthew; Manefield, Michael; Marquis, Christopher P. (2015-10-01). "Reductive Dehalogenases Come of Age in Biological Destruction of Organohalides". Trends in Biotechnology. 33 (10): 595–610. doi:10.1016/j.tibtech.2015.07.004. ISSN 0167-7799. PMID 26409778.
- ↑ Jugder, Bat-Erdene; Ertan, Haluk; Bohl, Susanne; Lee, Matthew; Marquis, Christopher P.; Manefield, Michael (2016). "Organohalide Respiring Bacteria and Reductive Dehalogenases: Key Tools in Organohalide Bioremediation". Frontiers in Microbiology. 7: 249. doi: 10.3389/fmicb.2016.00249 . ISSN 1664-302X. PMC 4771760 . PMID 26973626.