
The centromere is the specialized DNA sequence of a chromosome that links a pair of sister chromatids. During mitosis, spindle fibers attach to the centromere via the kinetochore. Centromeres were first thought to be genetic loci that direct the behavior of chromosomes.

In cell biology, mitosis is a part of the cell cycle in which replicated chromosomes are separated into two new nuclei. Cell division gives rise to genetically identical cells in which the total number of chromosomes is maintained. Therefore, mitosis is also known as equational division. In general, mitosis is preceded by the S stage of interphase and is often followed by telophase and cytokinesis; which divides the cytoplasm, organelles and cell membrane of one cell into two new cells containing roughly equal shares of these cellular components. The different stages of Mitosis altogether define the mitotic (M) phase of an animal cell cycle—the division of the mother cell into two daughter cells genetically identical to each other.
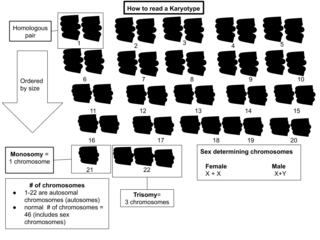
A karyotype is a preparation of the complete set of metaphase chromosomes in the cells of a species or in an individual organism, sorted by length, centromere location and other features. and for a test that detects this complement or counts the number of chromosomes. Karyotyping is the process by which a karyotype is prepared from photographs of chromosomes, in order to determine the chromosome complement of an individual, including the number of chromosomes and any abnormalities.

Aneuploidy is the presence of an abnormal number of chromosomes in a cell, for example a human cell having 45 or 47 chromosomes instead of the usual 46. It does not include a difference of one or more complete sets of chromosomes. A cell with any number of complete chromosome sets is called a euploid cell.

Cytogenetics is essentially a branch of genetics, but is also a part of cell biology/cytology, that is concerned with how the chromosomes relate to cell behaviour, particularly to their behaviour during mitosis and meiosis. Techniques used include karyotyping, analysis of G-banded chromosomes, other cytogenetic banding techniques, as well as molecular cytogenetics such as fluorescent in situ hybridization (FISH) and comparative genomic hybridization (CGH).

Nondisjunction is the failure of homologous chromosomes or sister chromatids to separate properly during cell division. There are three forms of nondisjunction: failure of a pair of homologous chromosomes to separate in meiosis I, failure of sister chromatids to separate during meiosis II, and failure of sister chromatids to separate during mitosis. Nondisjunction results in daughter cells with abnormal chromosome numbers (aneuploidy).

In genetics, chromosome translocation is a phenomenon that results in unusual rearrangement of chromosomes. This includes balanced and unbalanced translocation, with two main types: reciprocal-, and Robertsonian translocation. Reciprocal translocation is a chromosome abnormality caused by exchange of parts between non-homologous chromosomes. Two detached fragments of two different chromosomes are switched. Robertsonian translocation occurs when two non-homologous chromosomes get attached, meaning that given two healthy pairs of chromosomes, one of each pair "sticks" together.
A small supernumerary marker chromosome (sSMC) is an abnormal extra chromosome. It contains copies of parts of one or more normal chromosomes and like normal chromosomes is located in the cell's nucleus, is replicated and distributed into each daughter cell during cell division, and typically has genes which may be expressed. However, it may also be active in causing birth defects and neoplasms. The sSMC's small size makes it virtually undetectable using classical cytogenetic methods: the far larger DNA and gene content of the cell's normal chromosomes obscures those of the sSMC. Newer molecular techniques such as fluorescence in situ hybridization, next generation sequencing, comparative genomic hybridization, and highly specialized cytogenetic G banding analyses are required to study it. Using these methods, the DNA sequences and genes in sSMCs are identified and help define as well as explain any effect(s) it may have on individuals.
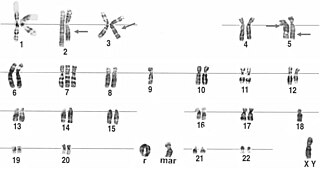
A ring chromosome is an aberrant chromosome whose ends have fused together to form a ring. Ring chromosomes were first discovered by Lilian Vaughan Morgan in 1926. A ring chromosome is denoted by the symbol r in human genetics and R in Drosophila genetics. Ring chromosomes may form in cells following genetic damage by mutagens like radiation, but they may also arise spontaneously during development.

An isochromosome is an unbalanced structural abnormality in which the arms of the chromosome are mirror images of each other. The chromosome consists of two copies of either the long (q) arm or the short (p) arm because isochromosome formation is equivalent to a simultaneous duplication and deletion of genetic material. Consequently, there is partial trisomy of the genes present in the isochromosome and partial monosomy of the genes in the lost arm.
Subtelomeres are segments of DNA between telomeric caps and chromatin.

Polysomy is a condition found in many species, including fungi, plants, insects, and mammals, in which an organism has at least one more chromosome than normal, i.e., there may be three or more copies of the chromosome rather than the expected two copies. Most eukaryotic species are diploid, meaning they have two sets of chromosomes, whereas prokaryotes are haploid, containing a single chromosome in each cell. Aneuploids possess chromosome numbers that are not exact multiples of the haploid number and polysomy is a type of aneuploidy. A karyotype is the set of chromosomes in an organism and the suffix -somy is used to name aneuploid karyotypes. This is not to be confused with the suffix -ploidy, referring to the number of complete sets of chromosomes.

Micronucleus is the name given to the small nucleus that forms whenever a chromosome or a fragment of a chromosome is not incorporated into one of the daughter nuclei during cell division. It usually is a sign of genotoxic events and chromosomal instability. Micronuclei are commonly seen in cancerous cells and may indicate genomic damage events that can increase the risk of developmental or degenerative diseases. Micronuclei form during anaphase from lagging acentric chromosome or chromatid fragments caused by incorrectly repaired or unrepaired DNA breaks or by nondisjunction of chromosomes. This incorrect segregation of chromosomes may result from hypomethylation of repeat sequences present in pericentromeric DNA, irregularities in kinetochore proteins or their assembly, dysfunctional spindle apparatus, or flawed anaphase checkpoint genes. Micronucleus can contribute to genome instability by promoting a catastrophic mutational event called chromothripsis. Many micronucleus assays have been developed to test for the presence of these structures and determine their frequency in cells exposed to certain chemicals or subjected to stressful conditions.
An acentric fragment is a segment of a chromosome that lacks a centromere.
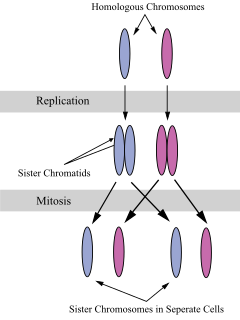
A sister chromatid refers to the identical copies (chromatids) formed by the DNA replication of a chromosome, with both copies joined together by a common centromere. In other words, a sister chromatid may also be said to be 'one-half' of the duplicated chromosome. A pair of sister chromatids is called a dyad. A full set of sister chromatids is created during the synthesis (S) phase of interphase, when all the chromosomes in a cell are replicated. The two sister chromatids are separated from each other into two different cells during mitosis or during the second division of meiosis.
A chromosome abnormality, chromosomal anomaly, chromosomal aberration, chromosomal mutation, or chromosomal disorder, is a missing, extra, or irregular portion of chromosomal DNA. These can occur in the form of numerical abnormalities, where there is an atypical number of chromosomes, or as structural abnormalities, where one or more individual chromosomes are altered. Chromosome mutation was formerly used in a strict sense to mean a change in a chromosomal segment, involving more than one gene. Chromosome anomalies usually occur when there is an error in cell division following meiosis or mitosis. Chromosome abnormalities may be detected or confirmed by comparing an individual's karyotype, or full set of chromosomes, to a typical karyotype for the species via genetic testing.
Inherited sterility in insects is induced by substerilizing doses of ionizing radiation. When partially sterile males mate with wild females, the radiation-induced deleterious effects are inherited by the F1 generation. As a result, egg hatch is reduced and the resulting offspring are both highly sterile and predominately male. Compared with the high radiation required to achieve full sterility in Lepidoptera, the lower dose of radiation used to induce F1 sterility increases the quality and competitiveness of the released insects as measured by improved dispersal after release, increased mating ability, and superior sperm competition.
Chromosomal instability (CIN) is a type of genomic instability in which chromosomes are unstable, such that either whole chromosomes or parts of chromosomes are duplicated or deleted. More specifically, CIN refers to the increase in rate of addition or loss of entire chromosomes or sections of them. The unequal distribution of DNA to daughter cells upon mitosis results in a failure to maintain euploidy leading to aneuploidy. In other words, the daughter cells do not have the same number of chromosomes as the cell they originated from. Chromosomal instability is the most common form of genetic instability and cause of aneuploidy.

Neocentromeres are new centromeres that form at a place on the chromosome that is usually not centromeric. They typically arise due to disruption of the normal centromere. These neocentromeres should not be confused with “knobs”, which were also described as “neocentromeres” in maize in the 1950s. Unlike most normal centromeres, neocentromeres do not contain satellite sequences that are highly repetitive but instead consist of unique sequences. Despite this, most neocentromeres are still able to carry out the functions of normal centromeres in regulating chromosome segregation and inheritance. This raises many questions on what is necessary versus what is sufficient for constituting a centromere.

The monocentric chromosome is a chromosome that has only one centromere in a chromosome and forms a narrow constriction.














