
A chromosome is a package of DNA with part or all of the genetic material of an organism. In most chromosomes, the very long thin DNA fibers are coated with nucleosome-forming packaging proteins; in eukaryotic cells the most important of these proteins are the histones. These proteins, aided by chaperone proteins, bind to and condense the DNA molecule to maintain its integrity. These chromosomes display a complex three-dimensional structure, which plays a significant role in transcriptional regulation.

The centromere links a pair of sister chromatids together during cell division. This constricted region of chromosome connects the sister chromatids, creating a short arm (p) and a long arm (q) on the chromatids. During mitosis, spindle fibers attach to the centromere via the kinetochore.

Meiosis is a special type of cell division of germ cells in sexually-reproducing organisms that produces the gametes, the sperm or egg cells. It involves two rounds of division that ultimately result in four cells, each with only one copy of each chromosome (haploid). Additionally, prior to the division, genetic material from the paternal and maternal copies of each chromosome is crossed over, creating new combinations of code on each chromosome. Later on, during fertilisation, the haploid cells produced by meiosis from a male and a female will fuse to create a zygote, a cell with two copies of each chromosome again.

Turner syndrome (TS), also known as 45,X, or 45,X0, is a genetic disorder in which a person's cells have only one X chromosome or are partially missing an X chromosome. Most people have two sex chromosomes. The chromosomal abnormality is often present in just some cells, in which case it is known as Turner syndrome with mosaicism. 45,X0 with mosaicism can occur in males or females, but Turner syndrome without mosaicism only occurs in females. Signs and symptoms vary among those affected. Often, a short and webbed neck, low-set ears, low hairline at the back of the neck, short stature, and swollen hands and feet are seen at birth. Typically, those affected do not develop menstrual periods or breasts without hormone treatment and are unable to have children without reproductive technology. Heart defects, diabetes, and hypothyroidism occur in the disorder more frequently than average. Most people with Turner syndrome have normal intelligence; however, many have problems with spatial visualization that may be needed in order to learn mathematics. Vision and hearing problems also occur more often than average.

Genetic recombination is the exchange of genetic material between different organisms which leads to production of offspring with combinations of traits that differ from those found in either parent. In eukaryotes, genetic recombination during meiosis can lead to a novel set of genetic information that can be further passed on from parents to offspring. Most recombination occurs naturally and can be classified into two types: (1) interchromosomal recombination, occurring through independent assortment of alleles whose loci are on different but homologous chromosomes ; & (2) intrachromosomal recombination, occurring through crossing over.

A chromatid is one half of a duplicated chromosome. Before replication, one chromosome is composed of one DNA molecule. In replication, the DNA molecule is copied, and the two molecules are known as chromatids. During the later stages of cell division these chromatids separate longitudinally to become individual chromosomes.

A pair of homologous chromosomes, or homologs, is a set of one maternal and one paternal chromosome that pair up with each other inside a cell during fertilization. Homologs have the same genes in the same loci, where they provide points along each chromosome that enable a pair of chromosomes to align correctly with each other before separating during meiosis. This is the basis for Mendelian inheritance, which characterizes inheritance patterns of genetic material from an organism to its offspring parent developmental cell at the given time and area.

Nondisjunction is the failure of homologous chromosomes or sister chromatids to separate properly during cell division (mitosis/meiosis). There are three forms of nondisjunction: failure of a pair of homologous chromosomes to separate in meiosis I, failure of sister chromatids to separate during meiosis II, and failure of sister chromatids to separate during mitosis. Nondisjunction results in daughter cells with abnormal chromosome numbers (aneuploidy).

In genetics, chromosome translocation is a phenomenon that results in unusual rearrangement of chromosomes. This includes balanced and unbalanced translocation, with two main types: reciprocal, and Robertsonian translocation. Reciprocal translocation is a chromosome abnormality caused by exchange of parts between non-homologous chromosomes. Two detached fragments of two different chromosomes are switched. Robertsonian translocation occurs when two non-homologous chromosomes get attached, meaning that given two healthy pairs of chromosomes, one of each pair "sticks" and blends together homogeneously.

Robertsonian translocation (ROB) is a chromosomal abnormality where the entire long arms of two different chromosomes become fused to each other. It is the most common form of chromosomal translocation in humans, affecting 1 out of every 1,000 babies born. It does not usually cause medical problems, though some people may produce gametes with an incorrect number of chromosomes, resulting in a risk of miscarriage. In rare cases this translocation results in Down syndrome and Patau syndrome. Robertsonian translocations result in a reduction in the number of chromosomes. A Robertsonian evolutionary fusion, which may have occurred in the common ancestor of humans and other great apes, is the reason humans have 46 chromosomes while all other primates have 48. Detailed DNA studies of chimpanzee, orangutan, gorilla and bonobo apes has determined that where human chromosome 2 is present in our DNA in all four great apes this is split into two separate chromosomes typically numbered 2a and 2b. Similarly, the fact that horses have 64 chromosomes and donkeys 62, and that they can still have common, albeit usually infertile, offspring, may be due to a Robertsonian evolutionary fusion at some point in the descent of today's donkeys from their common ancestor.

An inversion is a chromosome rearrangement in which a segment of a chromosome becomes inverted within its original position. An inversion occurs when a chromosome undergoes a two breaks within the chromosomal arm, and the segment between the two breaks inserts itself in the opposite direction in the same chromosome arm. The breakpoints of inversions often happen in regions of repetitive nucleotides, and the regions may be reused in other inversions. Chromosomal segments in inversions can be as small as 1 kilobases or as large as 100 megabases. The number of genes captured by an inversion can range from a handful of genes to hundreds of genes. Inversions can happen either through ectopic recombination between repetitive sequences, or through chromosomal breakage followed by non-homologous end joining.

A small supernumerary marker chromosome (sSMC) is an abnormal extra chromosome. It contains copies of parts of one or more normal chromosomes and like normal chromosomes is located in the cell's nucleus, is replicated and distributed into each daughter cell during cell division, and typically has genes which may be expressed. However, it may also be active in causing birth defects and neoplasms. The sSMC's small size makes it virtually undetectable using classical cytogenetic methods: the far larger DNA and gene content of the cell's normal chromosomes obscures those of the sSMC. Newer molecular techniques such as fluorescence in situ hybridization, next generation sequencing, comparative genomic hybridization, and highly specialized cytogenetic G banding analyses are required to study it. Using these methods, the DNA sequences and genes in sSMCs are identified and help define as well as explain any effect(s) it may have on individuals.
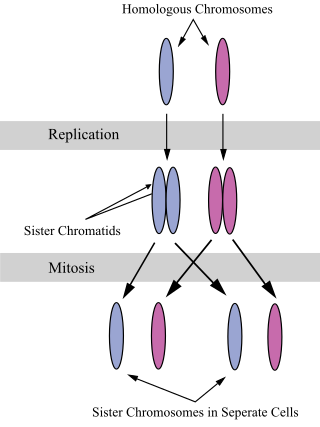
A sister chromatid refers to the identical copies (chromatids) formed by the DNA replication of a chromosome, with both copies joined together by a common centromere. In other words, a sister chromatid may also be said to be 'one-half' of the duplicated chromosome. A pair of sister chromatids is called a dyad. A full set of sister chromatids is created during the synthesis (S) phase of interphase, when all the chromosomes in a cell are replicated. The two sister chromatids are separated from each other into two different cells during mitosis or during the second division of meiosis.

Cohesin is a protein complex that mediates sister chromatid cohesion, homologous recombination, and DNA looping. Cohesin is formed of SMC3, SMC1, SCC1 and SCC3. Cohesin holds sister chromatids together after DNA replication until anaphase when removal of cohesin leads to separation of sister chromatids. The complex forms a ring-like structure and it is believed that sister chromatids are held together by entrapment inside the cohesin ring. Cohesin is a member of the SMC family of protein complexes which includes Condensin, MukBEF and SMC-ScpAB.
The Pallister–Killian syndrome (PKS), also termed tetrasomy 12p mosaicism or the Pallister mosaic aneuploidy syndrome, is an extremely rare and severe genetic disorder. PKS is due to the presence of an extra and abnormal chromosome termed a small supernumerary marker chromosome (sSMC). sSMCs contain copies of genetic material from parts of virtually any other chromosome and, depending on the genetic material they carry, can cause various genetic disorders and neoplasms. The sSMC in PKS consists of multiple copies of the short arm of chromosome 12. Consequently, the multiple copies of the genetic material in the sSMC plus the two copies of this genetic material in the two normal chromosome 12's are overexpressed and thereby cause the syndrome. Due to a form of genetic mosaicism, however, individuals with PKS differ in the tissue distributions of their sSMC and therefore show different syndrome-related birth defects and disease severities. For example, individuals with the sSMC in their heart tissue are likely to have cardiac structural abnormalities while those without this sSMC localization have a structurally normal heart.
A dicentric chromosome is an abnormal chromosome with two centromeres. It is formed through the fusion of two chromosome segments, each with a centromere, resulting in the loss of acentric fragments and the formation of dicentric fragments. The formation of dicentric chromosomes has been attributed to genetic processes, such as Robertsonian translocation and paracentric inversion. Dicentric chromosomes have important roles in the mitotic stability of chromosomes and the formation of pseudodicentric chromosomes. Their existence has been linked to certain natural phenomena such as irradiation and have been documented to underlie certain clinical syndromes, notably Kabuki syndrome. The formation of dicentric chromosomes and their implications on centromere function are studied in certain clinical cytogenetics laboratories.
A chromosomal abnormality, chromosomal anomaly, chromosomal aberration, chromosomal mutation, or chromosomal disorder is a missing, extra, or irregular portion of chromosomal DNA. These can occur in the form of numerical abnormalities, where there is an atypical number of chromosomes, or as structural abnormalities, where one or more individual chromosomes are altered. Chromosome mutation was formerly used in a strict sense to mean a change in a chromosomal segment, involving more than one gene. Chromosome anomalies usually occur when there is an error in cell division following meiosis or mitosis. Chromosome abnormalities may be detected or confirmed by comparing an individual's karyotype, or full set of chromosomes, to a typical karyotype for the species via genetic testing.
Tetrasomy 9p is a rare chromosomal disorder characterized by the presence of two extra copies of the short arm of chromosome 9, in addition to the usual two. Symptoms of tetrasomy 9p vary widely among affected individuals but typically include varying degrees of delayed growth, abnormal facial features and intellectual disability. Symptoms of the disorder are comparable to those of trisomy 9p.

Unequal crossing over is a type of gene duplication or deletion event that deletes a sequence in one strand and replaces it with a duplication from its sister chromatid in mitosis or from its homologous chromosome during meiosis. It is a type of chromosomal crossover between homologous sequences that are not paired precisely. Normally genes are responsible for occurrence of crossing over. It exchanges sequences of different links between chromosomes. Along with gene conversion, it is believed to be the main driver for the generation of gene duplications and is a source of mutation in the genome.

Neocentromeres are new centromeres that form at a place on the chromosome that is usually not centromeric. They typically arise due to disruption of the normal centromere. These neocentromeres should not be confused with “knobs”, which were also described as “neocentromeres” in maize in the 1950s. Unlike most normal centromeres, neocentromeres do not contain satellite sequences that are highly repetitive but instead consist of unique sequences. Despite this, most neocentromeres are still able to carry out the functions of normal centromeres in regulating chromosome segregation and inheritance. This raises many questions on what is necessary versus what is sufficient for constituting a centromere.

















