
Nucleic acids are biopolymers, macromolecules, essential to all known forms of life. They are composed of nucleotides, which are the monomer components: a 5-carbon sugar, a phosphate group and a nitrogenous base. The two main classes of nucleic acids are deoxyribonucleic acid (DNA) and ribonucleic acid (RNA). If the sugar is ribose, the polymer is RNA; if the sugar is deoxyribose, a version of ribose, the polymer is DNA.

Nucleotides are organic molecules composed of a nitrogenous base, a pentose sugar and a phosphate. They serve as monomeric units of the nucleic acid polymers – deoxyribonucleic acid (DNA) and ribonucleic acid (RNA), both of which are essential biomolecules within all life-forms on Earth. Nucleotides are obtained in the diet and are also synthesized from common nutrients by the liver.
DMT is an abbreviation of N,N-Dimethyltryptamine, a naturally occurring psychedelic compound.
Oligonucleotides are short DNA or RNA molecules, oligomers, that have a wide range of applications in genetic testing, research, and forensics. Commonly made in the laboratory by solid-phase chemical synthesis, these small bits of nucleic acids can be manufactured as single-stranded molecules with any user-specified sequence, and so are vital for artificial gene synthesis, polymerase chain reaction (PCR), DNA sequencing, molecular cloning and as molecular probes. In nature, oligonucleotides are usually found as small RNA molecules that function in the regulation of gene expression, or are degradation intermediates derived from the breakdown of larger nucleic acid molecules.

Guanosine-5'-triphosphate (GTP) is a purine nucleoside triphosphate. It is one of the building blocks needed for the synthesis of RNA during the transcription process. Its structure is similar to that of the guanosine nucleoside, the only difference being that nucleotides like GTP have phosphates on their ribose sugar. GTP has the guanine nucleobase attached to the 1' carbon of the ribose and it has the triphosphate moiety attached to ribose's 5' carbon.

Guanosine diphosphate, abbreviated GDP, is a nucleoside diphosphate. It is an ester of pyrophosphoric acid with the nucleoside guanosine. GDP consists of a pyrophosphate group, a pentose sugar ribose, and the nucleobase guanine.
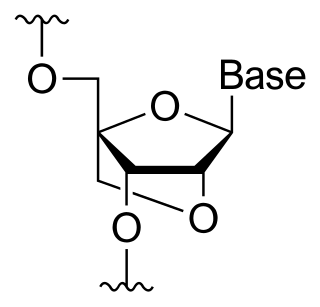
A locked nucleic acid (LNA), also known as bridged nucleic acid (BNA), and often referred to as inaccessible RNA, is a modified RNA nucleotide in which the ribose moiety is modified with an extra bridge connecting the 2' oxygen and 4' carbon. The bridge "locks" the ribose in the 3'-endo (North) conformation, which is often found in the A-form duplexes. This structure provides for increased stability against enzymatic degradation. LNA also offers improved specificity and affinity in base-pairing as a monomer or a constituent of an oligonucleotide. LNA nucleotides can be mixed with DNA or RNA residues in a oligonucleotide.

Fluorescein is an organic compound and dye based on the xanthene tricyclic structural motif, formally belonging to triarylmethine dyes family. It is available as a dark orange/red powder slightly soluble in water and alcohol. It is widely used as a fluorescent tracer for many applications.

A protecting group or protective group is introduced into a molecule by chemical modification of a functional group to obtain chemoselectivity in a subsequent chemical reaction. It plays an important role in multistep organic synthesis.
In chemistry, solid-phase synthesis is a method in which molecules are covalently bound on a solid support material and synthesised step-by-step in a single reaction vessel utilising selective protecting group chemistry. Benefits compared with normal synthesis in a liquid state include:

Uridine diphosphate, abbreviated UDP, is a nucleotide diphosphate. It is an ester of pyrophosphoric acid with the nucleoside uridine. UDP consists of the pyrophosphate group, the pentose sugar ribose, and the nucleobase uracil.
Mung bean nuclease is a nuclease derived from sprouts of the mung bean that removes nucleotides in a step-wise manner from single-stranded DNA molecules (ssDNA) and is used in biotechnological applications to remove such ssDNA from a mixture also containing double-stranded DNA (dsDNA). This enzyme is useful for transcript mapping, removal of single-stranded regions in DNA hybrids or single-stranded overhangs produced by restriction enzymes, etc. It has an activity similar to Nuclease S1, but it has higher specificity for single-stranded molecules.
Threose nucleic acid (TNA) is an artificial genetic polymer in which the natural five-carbon ribose sugar found in RNA has been replaced by an unnatural four-carbon threose sugar. Invented by Albert Eschenmoser as part of his quest to explore the chemical etiology of RNA, TNA has become an important synthetic genetic polymer (XNA) due to its ability to efficiently base pair with complementary sequences of DNA and RNA. However, unlike DNA and RNA, TNA is completely refractory to nuclease digestion, making it a promising nucleic acid analog for therapeutic and diagnostic applications.

Molecular beacons, or molecular beacon probes, are oligonucleotide hybridization probes that can report the presence of specific nucleic acids in homogenous solutions. Molecular beacons are hairpin-shaped molecules with an internally quenched fluorophore whose fluorescence is restored when they bind to a target nucleic acid sequence. This is a novel non-radioactive method for detecting specific sequences of nucleic acids. They are useful in situations where it is either not possible or desirable to isolate the probe-target hybrids from an excess of the hybridization probes.
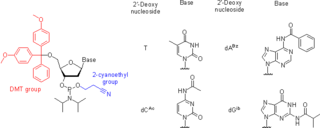
Nucleoside phosphoramidites are derivatives of natural or synthetic nucleosides. They are used to synthesize oligonucleotides, relatively short fragments of nucleic acid and their analogs. Nucleoside phosphoramidites were first introduced in 1981 by Beaucage and Caruthers. To avoid undesired side reactions, reactive hydroxy and exocyclic amino groups present in natural or synthetic nucleosides are appropriately protected. As long as a nucleoside analog contains at least one hydroxy group, the use of the appropriate protecting strategy allows one to convert that to the respective phosphoramidite and to incorporate the latter into synthetic nucleic acids. To be incorporated in the middle of an oligonucleotide chain using phosphoramidite strategy, the nucleoside analog must possess two hydroxy groups or, less often, a hydroxy group and another nucleophilic group (amino or mercapto). Examples include, but are not limited to, alternative nucleotides, LNA, morpholino, nucleosides modified at the 2'-position (OMe, protected NH2, F), nucleosides containing non-canonical bases (hypoxanthine and xanthine contained in natural nucleosides inosine and xanthosine, respectively, tricyclic bases such as G-clamp, etc.) or bases derivatized with a fluorescent group or a linker arm.
Oligonucleotide synthesis is the chemical synthesis of relatively short fragments of nucleic acids with defined chemical structure (sequence). The technique is extremely useful in current laboratory practice because it provides a rapid and inexpensive access to custom-made oligonucleotides of the desired sequence. Whereas enzymes synthesize DNA and RNA only in a 5' to 3' direction, chemical oligonucleotide synthesis does not have this limitation, although it is most often carried out in the opposite, 3' to 5' direction. Currently, the process is implemented as solid-phase synthesis using phosphoramidite method and phosphoramidite building blocks derived from protected 2'-deoxynucleosides, ribonucleosides, or chemically modified nucleosides, e.g. LNA or BNA.

Boranophosphates are salts with an anion consisting of borane (BH3) and phosphite groups. One of the simplest examples is [(CH3O)2OPBH3]−, prepared by base hydrolysis of the adduct of borane and trimethylphosphite.

Artificial gene synthesis, or simply gene synthesis, refers to a group of methods that are used in synthetic biology to construct and assemble genes from nucleotides de novo. Unlike DNA synthesis in living cells, artificial gene synthesis does not require template DNA, allowing virtually any DNA sequence to be synthesized in the laboratory. It comprises two main steps, the first of which is solid-phase DNA synthesis, sometimes known as DNA printing. This produces oligonucleotide fragments that are generally under 200 base pairs. The second step then involves connecting these oligonucleotide fragments using various DNA assembly methods. Because artificial gene synthesis does not require template DNA, it is theoretically possible to make a completely synthetic DNA molecule with no limits on the nucleotide sequence or size.

T7 DNA polymerase is an enzyme used during the DNA replication of the T7 bacteriophage. During this process, the DNA polymerase “reads” existing DNA strands and creates two new strands that match the existing ones. The T7 DNA polymerase requires a host factor, E. coli thioredoxin, in order to carry out its function. This helps stabilize the binding of the necessary protein to the primer-template to improve processivity by more than 100-fold, which is a feature unique to this enzyme. It is a member of the Family A DNA polymerases, which include E. coli DNA polymerase I and Taq DNA polymerase.
A bridged nucleic acid (BNA) is a modified RNA nucleotide. They are sometimes also referred to as constrained or inaccessible RNA molecules. BNA monomers can contain a five-membered, six-membered or even a seven-membered bridged structure with a "fixed" C3'-endo sugar puckering. The bridge is synthetically incorporated at the 2', 4'-position of the ribose to afford a 2', 4'-BNA monomer. The monomers can be incorporated into oligonucleotide polymeric structures using standard phosphoramidite chemistry. BNAs are structurally rigid oligo-nucleotides with increased binding affinities and stability.












