
Tandem mass spectrometry, also known as MS/MS or MS2, is a technique in instrumental analysis where two or more stages of analysis using one or more mass analyzer are performed with an additional reaction step in between these analyses to increase their abilities to analyse chemical samples. A common use of tandem MS is the analysis of biomolecules, such as proteins and peptides.
In biochemistry, biotinylation is the process of covalently attaching biotin to a protein, nucleic acid or other molecule. Biotinylation is rapid, specific and is unlikely to disturb the natural function of the molecule due to the small size of biotin. Biotin binds to streptavidin and avidin with an extremely high affinity, fast on-rate, and high specificity, and these interactions are exploited in many areas of biotechnology to isolate biotinylated molecules of interest. Biotin-binding to streptavidin and avidin is resistant to extremes of heat, pH and proteolysis, making capture of biotinylated molecules possible in a wide variety of environments. Also, multiple biotin molecules can be conjugated to a protein of interest, which allows binding of multiple streptavidin, avidin or neutravidin protein molecules and increases the sensitivity of detection of the protein of interest. There is a large number of biotinylation reagents available that exploit the wide range of possible labelling methods. Due to the strong affinity between biotin and streptavidin, the purification of biotinylated proteins has been a widely used approach to identify protein-protein interactions and post-translational events such as ubiquitylation in molecular biology.
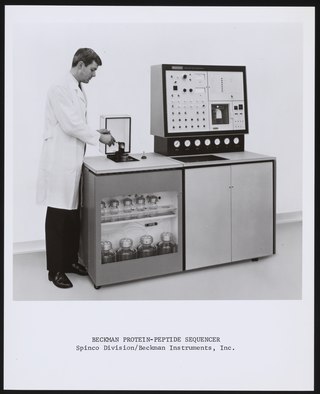
Protein sequencing is the practical process of determining the amino acid sequence of all or part of a protein or peptide. This may serve to identify the protein or characterize its post-translational modifications. Typically, partial sequencing of a protein provides sufficient information to identify it with reference to databases of protein sequences derived from the conceptual translation of genes.
In chemistry and biology a cross-link is a bond or a short sequence of bonds that links one polymer chain to another. These links may take the form of covalent bonds or ionic bonds and the polymers can be either synthetic polymers or natural polymers.

In organic chemistry, peptide synthesis is the production of peptides, compounds where multiple amino acids are linked via amide bonds, also known as peptide bonds. Peptides are chemically synthesized by the condensation reaction of the carboxyl group of one amino acid to the amino group of another. Protecting group strategies are usually necessary to prevent undesirable side reactions with the various amino acid side chains. Chemical peptide synthesis most commonly starts at the carboxyl end of the peptide (C-terminus), and proceeds toward the amino-terminus (N-terminus). Protein biosynthesis in living organisms occurs in the opposite direction.
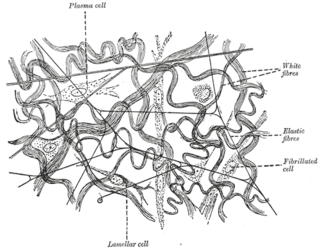
Elastic fibers are an essential component of the extracellular matrix composed of bundles of proteins (elastin) which are produced by a number of different cell types including fibroblasts, endothelial, smooth muscle, and airway epithelial cells. These fibers are able to stretch many times their length, and snap back to their original length when relaxed without loss of energy. Elastic fibers include elastin, elaunin and oxytalan.

2-Iodoacetamide (IAA) is an alkylating agent used for peptide mapping purposes. Its actions are similar to those of iodoacetate. It is commonly used to bind covalently with the thiol group of cysteine so the protein cannot form disulfide bonds. It is also used in ubiquitin studies as an inhibitor of deubiquitinase enzymes (DUBs) because it alkylates the cysteine residues at the DUB active site.
Cyanogen bromide is the inorganic compound with the formula (CN)Br or BrCN. It is a colorless solid that is widely used to modify biopolymers, fragment proteins and peptides, and synthesize other compounds. The compound is classified as a pseudohalogen.

Glucosepane is a lysine-arginine protein cross-linking product and advanced glycation end product (AGE) derived from D-glucose. It is an irreversible, covalent cross-link product that has been found to make intermolecular and intramolecular cross-links in the collagen of the extracellular matrix (ECM) and crystallin of the eyes. Covalent protein cross-links irreversibly link proteins together in the ECM of tissues. Glucosepane is present in human tissues at levels 10 to 1000 times higher than any other cross-linking AGE, and is currently considered to be the most important cross-linking AGE.
The Amadori rearrangement is an organic reaction describing the acid or base catalyzed isomerization or rearrangement reaction of the N-glycoside of an aldose or the glycosylamine to the corresponding 1-amino-1-deoxy-ketose. The reaction is important in carbohydrate chemistry, specifically the glycation of hemoglobin.
Carbamino refers to an adduct generated by the addition of carbon dioxide to the free amino group of an amino acid or a protein, such as hemoglobin forming carbaminohemoglobin.
Bioconjugation is a chemical strategy to form a stable covalent link between two molecules, at least one of which is a biomolecule.
Bissulfosuccinimidyl suberate (BS3) is a crosslinker used in biological research. It is a water-soluble version of disuccinimidyl suberate.

N-Hydroxysuccinimide (NHS) is an organic compound with the formula (CH2CO)2NOH. It is a white solid that is used as a reagent for preparing active esters in peptide synthesis. It can be synthesized by heating succinic anhydride with hydroxylamine or hydroxylamine hydrochloride.

Quantitative proteomics is an analytical chemistry technique for determining the amount of proteins in a sample. The methods for protein identification are identical to those used in general proteomics, but include quantification as an additional dimension. Rather than just providing lists of proteins identified in a certain sample, quantitative proteomics yields information about the physiological differences between two biological samples. For example, this approach can be used to compare samples from healthy and diseased patients. Quantitative proteomics is mainly performed by two-dimensional gel electrophoresis (2-DE), preparative native PAGE, or mass spectrometry (MS). However, a recent developed method of quantitative dot blot (QDB) analysis is able to measure both the absolute and relative quantity of an individual proteins in the sample in high throughput format, thus open a new direction for proteomic research. In contrast to 2-DE, which requires MS for the downstream protein identification, MS technology can identify and quantify the changes.

Isobaric tags for relative and absolute quantitation (iTRAQ) is an isobaric labeling method used in quantitative proteomics by tandem mass spectrometry to determine the amount of proteins from different sources in a single experiment. It uses stable isotope labeled molecules that can be covalent bonded to the N-terminus and side chain amines of proteins.

Isobaric labeling is a mass spectrometry strategy used in quantitative proteomics. Peptides or proteins are labeled with chemical groups that have identical mass (isobaric), but vary in terms of distribution of heavy isotopes in their structure. These tags, commonly referred to as tandem mass tags, are designed so that the mass tag is cleaved at a specific linker region upon high-energy CID (HCD) during tandem mass spectrometry yielding reporter ions of different masses. The most common isobaric tags are amine-reactive tags. However, tags that react with cysteine residues and carbonyl groups have also been described. These amine-reactive groups go through N-hydroxysuccinimide (NHS) reactions, which are based around three types of functional groups. Isobaric labeling methods include tandem mass tags (TMT), isobaric tags for relative and absolute quantification (iTRAQ), mass differential tags for absolute and relative quantification, and dimethyl labeling. TMTs and iTRAQ methods are most common and developed of these methods. Tandem mass tags have a mass reporter region, a cleavable linker region, a mass normalization region, and a protein reactive group and have the same total mass.
Terminal amine isotopic labeling of substrates (TAILS) is a method in quantitative proteomics that identifies the protein content of samples based on N-terminal fragments of each protein and detects differences in protein abundance among samples.

L-Photo-methionine is a photo-reactive amino acid derivative of L-methionine that was synthetically formed in 2005. Protein are long polymer chains of amino acids; which can range in various structures and sizes. Proteins can interact with each other and with these interactions, affects cellular interactions and pathways. Such interactions; in viral fusion and in growth-factor signaling looked promising for antiviral or anti-cancer drugs, so research must be done to understand the interactions. With that, research has begun to prove that proteins function in supramolecular complexes compared to isolated entities. So, scientists Monika Suchanek, Anna Radzikowski, and Christoph Thiele researched that the direct way to study these interactions in the natural environment better was to create a new way of photo-cross-linking proteins; which led to the synthesis of L-photo-methionine and in that same study, L-photo-leucine.

Ribonucleoprotein Networks Analyzed by Mutational Profiling (RNP-MaP) is a strategy for probing RNA-protein networks and protein binding sites at a nucleotide resolution. Information about RNP assembly and function can facilitate a better understanding of biological mechanisms. RNP-MaP uses NHS-diazirine (SDA), a hetero-bifunctional crosslinker, to freeze RNA-bound proteins in place. Once the RNA-protein crosslinks are formed, MaP reverse transcription is then conducted to reversely transcribe the protein-bound RNAs as well as introduce mutations at the site of RNA-protein crosslinks. Sequencing results of the cDNAs reveal information about both protein-RNA interaction networks and protein binding sites.













