Related Research Articles
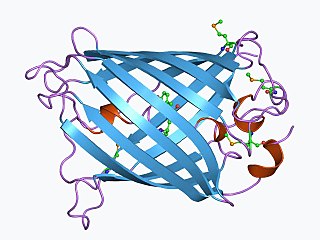
The green fluorescent protein (GFP) is a protein that exhibits green fluorescence when exposed to light in the blue to ultraviolet range. The label GFP traditionally refers to the protein first isolated from the jellyfish Aequorea victoria and is sometimes called avGFP. However, GFPs have been found in other organisms including corals, sea anemones, zoanithids, copepods and lancelets.

In molecular biology and biotechnology, a fluorescent tag, also known as a fluorescent label or fluorescent probe, is a molecule that is attached chemically to aid in the detection of a biomolecule such as a protein, antibody, or amino acid. Generally, fluorescent tagging, or labeling, uses a reactive derivative of a fluorescent molecule known as a fluorophore. The fluorophore selectively binds to a specific region or functional group on the target molecule and can be attached chemically or biologically. Various labeling techniques such as enzymatic labeling, protein labeling, and genetic labeling are widely utilized. Ethidium bromide, fluorescein and green fluorescent protein are common tags. The most commonly labelled molecules are antibodies, proteins, amino acids and peptides which are then used as specific probes for detection of a particular target.

A cloning vector is a small piece of DNA that can be stably maintained in an organism, and into which a foreign DNA fragment can be inserted for cloning purposes. The cloning vector may be DNA taken from a virus, the cell of a higher organism, or it may be the plasmid of a bacterium. The vector contains features that allow for the convenient insertion of a DNA fragment into the vector or its removal from the vector, for example through the presence of restriction sites. The vector and the foreign DNA may be treated with a restriction enzyme that cuts the DNA, and DNA fragments thus generated contain either blunt ends or overhangs known as sticky ends, and vector DNA and foreign DNA with compatible ends can then be joined by molecular ligation. After a DNA fragment has been cloned into a cloning vector, it may be further subcloned into another vector designed for more specific use.
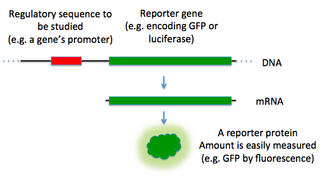
In molecular biology, a reporter gene is a gene that researchers attach to a regulatory sequence of another gene of interest in bacteria, cell culture, animals or plants. Such genes are called reporters because the characteristics they confer on organisms expressing them are easily identified and measured, or because they are selectable markers. Reporter genes are often used as an indication of whether a certain gene has been taken up by or expressed in the cell or organism population.

Aequorea victoria, also sometimes called the crystal jelly, is a bioluminescent hydrozoan jellyfish, or hydromedusa, that is found off the west coast of North America.

The pGLO plasmid is an engineered plasmid used in biotechnology as a vector for creating genetically modified organisms. The plasmid contains several reporter genes, most notably the green fluorescent protein (GFP) and the ampicillin resistance gene. GFP was isolated from the jelly fish Aequorea victoria. Because it shares a bidirectional promoter with a gene for metabolizing arabinose, the GFP gene is expressed in the presence of arabinose, which makes the transgenic organism express its fluorescence under UV light. GFP can be induced in bacteria containing the pGLO plasmid by growing them on +arabinose plates. pGLO is made by Bio-Rad Laboratories.

Aequorin is a calcium-activated photoprotein isolated from the hydrozoan Aequorea victoria. Its bioluminescence was studied decades before the protein was isolated from the animal by Osamu Shimomura in 1962. In the animal, the protein occurs together with the green fluorescent protein to produce green light by resonant energy transfer, while aequorin by itself generates blue light.
The GFP-cDNA project documents the localisation of proteins to subcellular compartments of the eukaryotic cell applying fluorescence microscopy. Experimental data are complemented with bioinformatic analyses and published online in a database. A search function allows the finding of proteins containing features or motifs of particular interest. The project is a collaboration of the research groups of Rainer Pepperkok at the European Molecular Biology Laboratory (EMBL) and Stefan Wiemann at the German Cancer Research Centre (DKFZ).
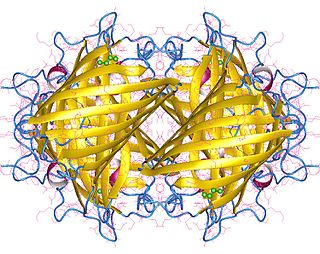
Yellow fluorescent protein (YFP) is a genetic mutant of green fluorescent protein (GFP) originally derived from the jellyfish Aequorea victoria. Its excitation peak is 513 nm and its emission peak is 527 nm. Like the parent GFP, YFP is a useful tool in cell and molecular biology because the excitation and emission peaks of YFP are distinguishable from GFP which allows for the study of multiple processes/proteins within the same experiment.
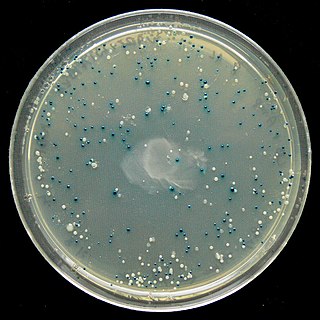
The blue–white screen is a screening technique that allows for the rapid and convenient detection of recombinant bacteria in vector-based molecular cloning experiments. This method of screening is usually performed using a suitable bacterial strain, but other organisms such as yeast may also be used. DNA of transformation is ligated into a vector. The vector is then inserted into a competent host cell viable for transformation, which are then grown in the presence of X-gal. Cells transformed with vectors containing recombinant DNA will produce white colonies; cells transformed with non-recombinant plasmids grow into blue colonies.

Roger Yonchien Tsien was an American biochemist. He was a professor of chemistry and biochemistry at the University of California, San Diego and was awarded the Nobel Prize in Chemistry in 2008 for his discovery and development of the green fluorescent protein, in collaboration with organic chemist Osamu Shimomura and neurobiologist Martin Chalfie. Tsien was also a pioneer of calcium imaging.

Aequorea forskalea is a species of hydrozoan in the family Aequoreidae. Discovered in 1810 by Péron and Lesueur, A. forskalea was initially found in coastal to offshore waters of the Mediterranean Sea. This species is commonly referred to as the many-ribbed jellyfish. The species is often mixed up with some other members of the genus due to some similarities including the capability of bioluminescence.
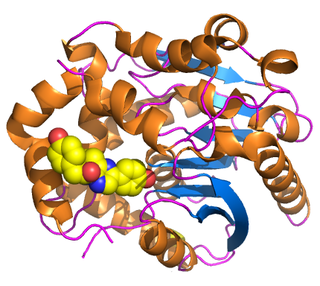
Renilla-luciferin 2-monooxygenase, Renilla luciferase, or RLuc, is a bioluminescent enzyme found in Renilla reniformis, belonging to a group of coelenterazine luciferases. Of this group of enzymes, the luciferase from Renilla reniformis has been the most extensively studied, and due to its bioluminescence requiring only molecular oxygen, has a wide range of applications, with uses as a reporter gene probe in cell culture, in vivo imaging, and various other areas of biological research. Recently, chimeras of RLuc have been developed and demonstrated to be the brightest luminescent proteins to date, and have proved effective in both noninvasive single-cell and whole body imaging.

Bioreporters are intact, living microbial cells that have been genetically engineered to produce a measurable signal in response to a specific chemical or physical agent in their environment. Bioreporters contain two essential genetic elements, a promoter gene and a reporter gene. The promoter gene is turned on (transcribed) when the target agent is present in the cell’s environment. The promoter gene in a normal bacterial cell is linked to other genes that are then likewise transcribed and then translated into proteins that help the cell in either combating or adapting to the agent to which it has been exposed. In the case of a bioreporter, these genes, or portions thereof, have been removed and replaced with a reporter gene. As a result, turning on the promoter gene also turns on the reporter gene, leading to the production of reporter proteins that output a detectable signal. The presence of a signal indicates that the bioreporter has sensed a particular agent in its environment.

Osamu Shimomura was a Japanese organic chemist and marine biologist, and professor emeritus at Marine Biological Laboratory (MBL) in Woods Hole, Massachusetts and Boston University School of Medicine. He was awarded the Nobel Prize in Chemistry in 2008 for the discovery and development of green fluorescent protein (GFP) with two American scientists: Martin Chalfie of Columbia University and Roger Tsien of the University of California-San Diego.

Martin Lee Chalfie is an American scientist. He is University Professor at Columbia University. He shared the 2008 Nobel Prize in Chemistry along with Osamu Shimomura and Roger Y. Tsien "for the discovery and development of the green fluorescent protein, GFP". He holds a PhD in neurobiology from Harvard University.
Paul Brehm is a researcher at the Vollum Institute at Oregon Health and Science University. It was during a seminar by Brehm that Martin Chalfie became inspired to work on Green fluorescent protein for which Chalfie shared the Nobel Prize in Chemistry in 2008.

Fluorescence is used in the life sciences generally as a non-destructive way of tracking or analysing biological molecules. Some proteins or small molecules in cells are naturally fluorescent, which is called intrinsic fluorescence or autofluorescence. Alternatively, specific or general proteins, nucleic acids, lipids or small molecules can be "labelled" with an extrinsic fluorophore, a fluorescent dye which can be a small molecule, protein or quantum dot. Several techniques exist to exploit additional properties of fluorophores, such as fluorescence resonance energy transfer, where the energy is passed non-radiatively to a particular neighbouring dye, allowing proximity or protein activation to be detected; another is the change in properties, such as intensity, of certain dyes depending on their environment allowing their use in structural studies.
mCherry is a member of the mFruits family of monomeric red fluorescent proteins (mRFPs). As an RFP, mCherry was derived from DsRed of Discosoma sea anemones, unlike green fluorescent proteins (GFPs) which are often derived from Aequorea victoria jellyfish. Fluorescent proteins are used to tag components in cells so that they can be studied using fluorescence spectroscopy and fluorescence microscopy. mCherry absorbs light between 540 and 590 nm and emits light in the range of 550-650 nm. mCherry belongs to the group of fluorescent protein chromophores used as instruments to visualize genes and analyze their functions in experiments. Genome editing has been improved greatly through the precise insertion of these fluorescent protein tags into the genetic material of many diverse organisms. Most comparisons between the brightness and photostability of different fluorescent proteins have been made in vitro, removed from biological variables that affect protein performance in cells or organisms. It is hard to perfectly simulate cellular environments in vitro, and the difference in environment could have an effect on the brightness and photostability.
Tulle Inger Hazelrigg is an American biologist who is Professor of Cell Biology at Columbia University. Her research considers the propagation and differentiation of germ cells. Hazelrigg was the first to attach green fluorescent protein to other proteins, which changed the way biological research could be conducted.
References
- 1 2 Prasher, D C; R O McCann; M Longiaru; M J Cormier (1987-03-10). "Sequence comparisons of complementary DNAs encoding aequorin isotypes". Biochemistry. 26 (5): 1326–1332. doi:10.1021/bi00379a019. ISSN 0006-2960. PMID 2882777.
- 1 2 Prasher, D C; V K Eckenrode; W W Ward; F G Prendergast; M J Cormier (1992-02-15). "Primary structure of the Aequorea victoria green-fluorescent protein". Gene. 111 (2): 229–233. doi:10.1016/0378-1119(92)90691-H. ISSN 0378-1119. PMID 1347277.
- ↑ Prasher, D C (August 1995). "Using GFP to see the light". Trends in Genetics. 11 (8): 320–323. doi:10.1016/s0168-9525(00)89090-3. ISSN 0168-9525. PMID 8585130.
- ↑ Herper, Matthew (2008-10-08). "Biotech's Glowing Breakthrough Wins Nobel Prize". Forbes. Archived from the original on October 11, 2008. Retrieved 2008-10-13.
- ↑ Chang, Kenneth (2008-10-09). "Three Chemists Win Nobel Prize". New York Times. Retrieved 2008-10-11.
- 1 2 3 Charles, Dan (2008-10-09). "Glowing Gene's Discoverer Left Out Of Nobel Prize". Morning Edition (National Public Radio [NPR]). Retrieved 2008-10-05.
- ↑ Saey, Tina Hesman (2008-10-25). "Nobel Prize in Chemistry Commends Finding and Use of Green Fluorescent Protein". Science News . Retrieved 2008-10-11.
- 1 2 Chalfie, M; Y Tu; G Euskirchen; W W Ward; D C Prasher (1994-02-11). "Green fluorescent protein as a marker for gene expression". Science. 263 (5148): 802–805. doi:10.1126/science.8303295. ISSN 0036-8075. PMID 8303295.
- 1 2 Haseloff, J; K R Siemering; D C Prasher; S Hodge (1997-03-18). "Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly". Proceedings of the National Academy of Sciences of the United States of America. 94 (6): 2122–2127. doi: 10.1073/pnas.94.6.2122 . ISSN 0027-8424. PMC 20051 . PMID 9122158.
- ↑ Chalfie, Martin; Tu, Yuan; Euskirchen, Ghia; Ward, William W.; Prasher, Douglas C. (1994-02-11). "Green Fluorescent Protein as a Marker for Gene Expression" . Science. 263 (5148): 802–805. doi:10.1126/science.8303295. ISSN 0036-8075.
- 1 2 3 "Van driver's work in Mass. aided Nobel winners". Boston Globe. 2008-10-11. Archived from the original on October 14, 2008. Retrieved 2014-03-17.
- 1 2 3 Doyle, Steve (2008-10-10). "Local biochemist had hand in Nobel". Huntsville Times. Archived from the original on 2011-06-09. Retrieved 2008-10-11.
- ↑ "The Nobel Prize in Chemistry 2008" (Press release). Royal Swedish Academy of Sciences. 8 October 2008. Retrieved 2008-10-11.
- 1 2 Gouveia, Aaron (2008-10-11). "Shuttle driver reflects on Nobel snub". Cape Cod Times. Archived from the original on 2008-10-11. Retrieved 2008-10-11.
- 1 2 Moret, Jim (2008-10-14). "Genius Behind the Wheel - Why is a genius scientist driving a bus?". Inside Edition . Archived from the original on 2012-06-20. Retrieved 2017-10-07.
- ↑ Sherwell, Philip (2008-10-11). "The scientist, the jellyfish protein, and the Nobel prize that got away". The Daily Telegraph. Retrieved 2008-10-11.
- ↑ Doyle, Steve (2008-12-04). "Van driver to attend Nobel ceremony". Huntsville Times. Archived from the original on 2011-06-09. Retrieved 2008-12-11.
- ↑ Roop, Lee (2008-12-18). "'Magical' Nobel trip could lead to new opportunities". Huntsville Times. Archived from the original on 2011-06-09. Retrieved 2009-02-04.
- ↑ Tsien Lab Staff
- ↑ Chang, Kenneth (4 September 2016). "Roger Y. Tsien, Nobel Winner for Use of Glowing Proteins, Dies at 64". New York Times. Retrieved 11 April 2023.