Enoyl-CoA-(∆) isomerase (EC 5.3.3.8, also known as dodecenoyl-CoA- isomerase, 3,2-trans-enoyl-CoA isomerase, ∆3 ,∆2 -enoyl-CoA isomerase, or acetylene-allene isomerase, is an enzyme that catalyzes the conversion of cis- or trans-double bonds of coenzyme A bound fatty acids at gamma-carbon to trans double bonds at beta-carbon as below:

Mitochondrial 5-demethoxyubiquinone hydroxylase, also known as coenzyme Q7, hydroxylase, is an enzyme that in humans is encoded by the COQ7 gene. The clk-1 (clock-1) gene encodes this protein that is necessary for ubiquinone biosynthesis in the worm Caenorhabditis elegans and other eukaryotes. The mouse version of the gene is called mclk-1 and the human, fruit fly and yeast homolog COQ7.
Mitochondrial trifunctional protein (MTP) is a protein attached to the inner mitochondrial membrane which catalyzes three out of the four steps in beta oxidation. MTP is a hetero-octamer composed of four alpha and four beta subunits:

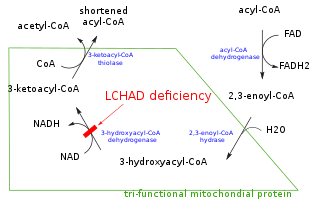
Trifunctional enzyme subunit alpha, mitochondrial also known as hydroxyacyl-CoA dehydrogenase/3-ketoacyl-CoA thiolase/enoyl-CoA hydratase, alpha subunit is a protein that in humans is encoded by the HADHA gene. Mutations in HADHA have been associated with trifunctional protein deficiency or long-chain 3-hydroxyacyl-coenzyme A dehydrogenase deficiency.

Trifunctional enzyme subunit beta, mitochondrial (TP-beta) also known as 3-ketoacyl-CoA thiolase, acetyl-CoA acyltransferase, or beta-ketothiolase is an enzyme that in humans is encoded by the HADHB gene.

2,4 Dienoyl-CoA reductase also known as DECR1 is an enzyme which in humans is encoded by the DECR1 gene which resides on chromosome 8. This enzyme catalyzes the following reactions
The crotonase family comprises mechanistically diverse proteins that share a conserved trimeric quaternary structure, the core of which consists of 4 turns of a (beta/beta/alpha)n superhelix.

3-Methylglutaconyl-CoA hydratase, also known as MG-CoA hydratase and AUH, is an enzyme encoded by the AUH gene on chromosome 19. It is a member of the enoyl-CoA hydratase/isomerase superfamily, but it is the only member of that family that is able to bind to RNA. Not only does it bind to RNA, AUH has also been observed to be involved in the metabolic enzymatic activity, making it a dual-role protein. Mutations of this gene have been found to cause a disease called 3-Methylglutaconic Acuduria Type 1.

D-bifunctional protein (DBP), also known as peroxisomal multifunctional enzyme type 2 (MFP-2), as well as 17β-hydroxysteroid dehydrogenase type IV is a protein that in humans is encoded by the HSD17B4 gene. It's an alcohol oxidoreductase, specifically 17β-Hydroxysteroid dehydrogenase. It is involved in fatty acid β-oxidation and steroid metabolism.

Peroxisomal acyl-coenzyme A oxidase 1 is an enzyme that in humans is encoded by the ACOX1 gene.

Peroxisomal 3,2-trans-enoyl-CoA isomerase is an enzyme that in humans is encoded by the PECI gene.

Acyl-CoA thioesterase 2, also known as ACOT2, is an enzyme which in humans is encoded by the ACOT2 gene.

NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 5 is an enzyme that in humans is encoded by the NDUFA5 gene. The NDUFA5 protein is a subunit of NADH dehydrogenase (ubiquinone), which is located in the mitochondrial inner membrane and is the largest of the five complexes of the electron transport chain.

Very long-chain acyl-CoA synthetase is an enzyme that in humans is encoded by the SLC27A2 gene.

Glycine-N-acyltransferase, also known as GLYAT, is an enzyme which in humans is encoded by the GLYAT gene.

Acyl-coenzyme A thioesterase 11 also known as StAR-related lipid transfer protein 14 (STARD14) is an enzyme that in humans is encoded by the ACOT11 gene. This gene encodes a protein with acyl-CoA thioesterase activity towards medium (C12) and long-chain (C18) fatty acyl-CoA substrates which relies on its StAR-related lipid transfer domain. Expression of a similar murine protein in brown adipose tissue is induced by cold exposure and repressed by warmth. Expression of the mouse protein has been associated with obesity, with higher expression found in obesity-resistant mice compared with obesity-prone mice. Alternative splicing results in two transcript variants encoding different isoforms.

Trans-2-enoyl-CoA reductase, mitochondrial is an enzyme that in humans is encoded by the MECR gene.

Delta(3,5)-Delta(2,4)-dienoyl-CoA isomerase, mitochondrial is an enzyme that in humans is encoded by the ECH1 gene.

Peroxisomal trans-2-enoyl-CoA reductase is an enzyme that in humans is encoded by the PECR gene.

Hydroxyacyl-Coenzyme A dehydrogenase (HADH) is an enzyme which in humans is encoded by the HADH gene.