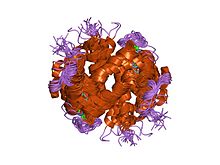
Actin is a family of globular multi-functional proteins that form microfilaments. It is found in essentially all eukaryotic cells, where it may be present at a concentration of over 100 μM; its mass is roughly 42-kDa, with a diameter of 4 to 7 nm.

Intermediate filaments (IFs) are cytoskeletal structural components found in the cells of vertebrates, and many invertebrates. Homologues of the IF protein have been noted in an invertebrate, the cephalochordate Branchiostoma.
In biology and biochemistry, protease inhibitors, or antiproteases, are molecules that inhibit the function of proteases. Many naturally occurring protease inhibitors are proteins.

Ku is a dimeric protein complex that binds to DNA double-strand break ends and is required for the non-homologous end joining (NHEJ) pathway of DNA repair. Ku is evolutionarily conserved from bacteria to humans. The ancestral bacterial Ku is a homodimer. Eukaryotic Ku is a heterodimer of two polypeptides, Ku70 (XRCC6) and Ku80 (XRCC5), so named because the molecular weight of the human Ku proteins is around 70 kDa and 80 kDa. The two Ku subunits form a basket-shaped structure that threads onto the DNA end. Once bound, Ku can slide down the DNA strand, allowing more Ku molecules to thread onto the end. In higher eukaryotes, Ku forms a complex with the DNA-dependent protein kinase catalytic subunit (DNA-PKcs) to form the full DNA-dependent protein kinase, DNA-PK. Ku is thought to function as a molecular scaffold to which other proteins involved in NHEJ can bind, orienting the double-strand break for ligation.
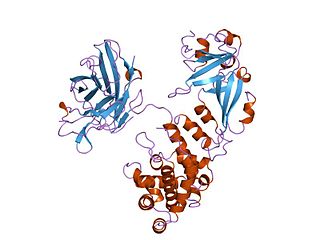
Diphtheria toxin is an exotoxin secreted by Corynebacterium, the pathogenic bacterium that causes diphtheria. The toxin gene is encoded by a prophage. The toxin causes the disease in humans by gaining entry into the cell cytoplasm and inhibiting protein synthesis.

A leucine-rich repeat (LRR) is a protein structural motif that forms an α/β horseshoe fold. It is composed of repeating 20–30 amino acid stretches that are unusually rich in the hydrophobic amino acid leucine. These tandem repeats commonly fold together to form a solenoid protein domain, termed leucine-rich repeat domain. Typically, each repeat unit has beta strand-turn-alpha helix structure, and the assembled domain, composed of many such repeats, has a horseshoe shape with an interior parallel beta sheet and an exterior array of helices. One face of the beta sheet and one side of the helix array are exposed to solvent and are therefore dominated by hydrophilic residues. The region between the helices and sheets is the protein's hydrophobic core and is tightly sterically packed with leucine residues.
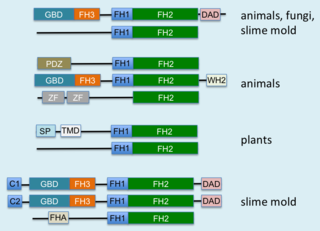
Formins are a group of proteins that are involved in the polymerization of actin and associate with the fast-growing end of actin filaments. Most formins are Rho-GTPase effector proteins. Formins regulate the actin and microtubule cytoskeleton and are involved in various cellular functions such as cell polarity, cytokinesis, cell migration and SRF transcriptional activity. Formins are multidomain proteins that interact with diverse signalling molecules and cytoskeletal proteins, although some formins have been assigned functions within the nucleus.

F-actin-capping protein subunit alpha-1 is a protein that in humans is encoded by the CAPZA1 gene.

The outer and inner segments of vertebrate retina rod photoreceptor cells contain phosducin, a soluble phosphoprotein that complexes with the beta/gamma-subunits of the guanosine triphosphate-binding protein, transducin. Light-induced changes in cyclic nucleotide levels modulate the phosphorylation of phosducin by protein kinase A. The protein is thought to participate in the regulation of visual phototransduction or in the integration of photoreceptor metabolism. Similar proteins have been isolated from the pineal gland and it is believed that the function of the protein is the same in both retina and pineal gland.

F-actin-capping protein subunit alpha-2 also known as CapZ-alpha2 is a protein that in humans is encoded by the CAPZA2 gene.

F-actin-capping protein subunit beta, also known as CapZβ is a protein that in humans is encoded by the CAPZB gene. CapZβ functions to cap actin filaments at barbed ends in muscle and other tissues.

In molecular biology, Phosphotyrosine-binding domains are protein domains which bind to phosphotyrosine.
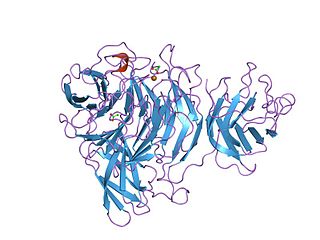
The Kelch motif is a region of protein sequence found widely in proteins from bacteria and eukaryotes. This sequence motif is composed of about 50 amino acid residues which form a structure of a four stranded beta-sheet "blade". This sequence motif is found in between five and eight tandem copies per protein which fold together to form a larger circular solenoid structure called a beta-propeller domain.
Vinculin is a eukaryotic protein that seems to be involved in the attachment of the actin-based microfilaments to the plasma membrane. Vinculin is located at the cytoplasmic side of focal contacts or adhesion plaques. In addition to actin, vinculin interacts with other structural proteins such as talin and alpha-actinins.
WH1 domain is an evolutionary conserved protein domain. Therefore, it has an important function.

In molecular biology, the cyclase-associated protein family (CAP) is a family of highly conserved actin-binding proteins present in a wide range of organisms including yeast, flies, plants, and mammals. CAPs are multifunctional proteins that contain several structural domains. CAP is involved in species-specific signalling pathways. In Drosophila, CAP functions in Hedgehog-mediated eye development and in establishing oocyte polarity. In Dictyostelium discoideum, CAP is involved in microfilament reorganisation near the plasma membrane in a PIP2-regulated manner and is required to perpetuate the cAMP relay signal to organise fruitbody formation. In plants, CAP is involved in plant signalling pathways required for co-ordinated organ expansion. In yeast, CAP is involved in adenylate cyclase activation, as well as in vesicle trafficking and endocytosis. In both yeast and mammals, CAPs appear to be involved in recycling G-actin monomers from ADF/cofilins for subsequent rounds of filament assembly. In mammals, there are two different CAPs that share 64% amino acid identity.

In molecular biology, a carbohydrate-binding module (CBM) is a protein domain found in carbohydrate-active enzymes. The majority of these domains have carbohydrate-binding activity. Some of these domains are found on cellulosomal scaffoldin proteins. CBMs were previously known as cellulose-binding domains. CBMs are classified into numerous families, based on amino acid sequence similarity. There are currently 64 families of CBM in the CAZy database.

In molecular biology, ADF-H domain is an approximately 150 amino acid motif that is present in three phylogenetically distinct classes of eukaryotic actin-binding proteins.
In molecular biology, the FERM domain is a widespread protein module involved in localising proteins to the plasma membrane. FERM domains are found in a number of cytoskeletal-associated proteins that associate with various proteins at the interface between the plasma membrane and the cytoskeleton. The FERM domain is located at the N terminus in the majority of proteins in which it is found.