Related Research Articles

In biology, histones are highly basic proteins abundant in lysine and arginine residues that are found in eukaryotic cell nuclei. They act as spools around which DNA winds to create structural units called nucleosomes. Nucleosomes in turn are wrapped into 30-nanometer fibers that form tightly packed chromatin. Histones prevent DNA from becoming tangled and protect it from DNA damage. In addition, histones play important roles in gene regulation and DNA replication. Without histones, unwound DNA in chromosomes would be very long. For example, each human cell has about 1.8 meters of DNA if completely stretched out, however when wound about histones, this length is reduced to about 90 micrometers (0.09 mm) of 30 nm diameter chromatin fibers.
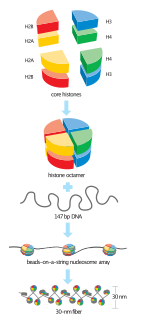
A nucleosome is the basic structural unit of DNA packaging in eukaryotes. The structure of a nucleosome consists of a segment of DNA wound around eight histone proteins and resembles thread wrapped around a spool. The nucleosome is the fundamental subunit of chromatin. Each nucleosome is composed of a little less than two turns of DNA wrapped around a set of eight proteins called histones, which are known as a histone octamer. Each histone octamer is composed of two copies each of the histone proteins H2A, H2B, H3, and H4.

Histone acetyltransferases (HATs) are enzymes that acetylate conserved lysine amino acids on histone proteins by transferring an acetyl group from acetyl-CoA to form ε-N-acetyllysine. DNA is wrapped around histones, and, by transferring an acetyl group to the histones, genes can be turned on and off. In general, histone acetylation increases gene expression.

A histone octamer is the eight protein complex found at the center of a nucleosome core particle. It consists of two copies of each of the four core histone proteins. The octamer assembles when a tetramer, containing two copies of both H3 and H4, complexes with two H2A/H2B dimers. Each histone has both an N-terminal tail and a C-terminal histone-fold. Both of these key components interact with DNA in their own way through a series of weak interactions, including hydrogen bonds and salt bridges. These interactions keep the DNA and histone octamer loosely associated and ultimately allow the two to re-position or separate entirely.
RNA polymerase 1 is, in higher eukaryotes, the polymerase that only transcribes ribosomal RNA, a type of RNA that accounts for over 50% of the total RNA synthesized in a cell.

RNA polymerase II is a multiprotein complex that transcribes DNA into precursors of messenger RNA (mRNA) and most small nuclear RNA (snRNA) and microRNA. It is one of the three RNAP enzymes found in the nucleus of eukaryotic cells. A 550 kDa complex of 12 subunits, RNAP II is the most studied type of RNA polymerase. A wide range of transcription factors are required for it to bind to upstream gene promoters and begin transcription.

General transcription factors (GTFs), also known as basal transcriptional factors, are a class of protein transcription factors that bind to specific sites (promoter) on DNA to activate transcription of genetic information from DNA to messenger RNA. GTFs, RNA polymerase, and the mediator constitute the basic transcriptional apparatus that first bind to the promoter, then start transcription. GTFs are also intimately involved in the process of gene regulation, and most are required for life.
Histone H2B is one of the 4 main histone proteins involved in the structure of chromatin in eukaryotic cells. Featuring a main globular domain and long N-terminal and C-terminal tails, H2B is involved with the structure of the nucleosomes.

Eukaryotic DNA replication is a conserved mechanism that restricts DNA replication to once per cell cycle. Eukaryotic DNA replication of chromosomal DNA is central for the duplication of a cell and is necessary for the maintenance of the eukaryotic genome.

Eukaryotic transcription is the elaborate process that eukaryotic cells use to copy genetic information stored in DNA into units of transportable complementary RNA replica. Gene transcription occurs in both eukaryotic and prokaryotic cells. Unlike prokaryotic RNA polymerase that initiates the transcription of all different types of RNA, RNA polymerase in eukaryotes comes in three variations, each translating a different type of gene. A eukaryotic cell has a nucleus that separates the processes of transcription and translation. Eukaryotic transcription occurs within the nucleus where DNA is packaged into nucleosomes and higher order chromatin structures. The complexity of the eukaryotic genome necessitates a great variety and complexity of gene expression control.
Chromatin remodeling is the dynamic modification of chromatin architecture to allow access of condensed genomic DNA to the regulatory transcription machinery proteins, and thereby control gene expression. Such remodeling is principally carried out by 1) covalent histone modifications by specific enzymes, e.g., histone acetyltransferases (HATs), deacetylases, methyltransferases, and kinases, and 2) ATP-dependent chromatin remodeling complexes which either move, eject or restructure nucleosomes. Besides actively regulating gene expression, dynamic remodeling of chromatin imparts an epigenetic regulatory role in several key biological processes, egg cells DNA replication and repair; apoptosis; chromosome segregation as well as development and pluripotency. Aberrations in chromatin remodeling proteins are found to be associated with human diseases, including cancer. Targeting chromatin remodeling pathways is currently evolving as a major therapeutic strategy in the treatment of several cancers.
DNA-directed RNA polymerase II subunit RPB1, also known as RPB1, is an enzyme that in humans is encoded by the POLR2A gene.

FACT complex subunit SSRP1 also known as structure specific recognition protein 1 is a protein that in humans is encoded by the SSRP1 gene.

FACT complex subunit SPT16 is a protein that in humans is encoded by the SUPT16H gene.

Mediator of RNA polymerase II transcription subunit 21 is an enzyme that in humans is encoded by the MED21 gene.

Transcription initiation protein SPT3 homolog is a protein that in humans is encoded by the SUPT3H gene.
RNA polymerase II holoenzyme is a form of eukaryotic RNA polymerase II that is recruited to the promoters of protein-coding genes in living cells. It consists of RNA polymerase II, a subset of general transcription factors, and regulatory proteins known as SRB proteins.
Cryptic unstable transcripts (CUTs) are a subset of non-coding RNAs (ncRNAs) that are produced from intergenic and intragenic regions. CUTs were first observed in S. cerevisiae yeast models and are found in most eukaryotes. Some basic characteristics of CUTs include a length of around 200–800 base pairs, a 5' cap, poly-adenylated tail, and rapid degradation due to the combined activity of poly-adenylating polymerases and exosome complexes. CUT transcription occurs through RNA Polymerase II and initiates from nucleosome-depleted regions, often in an antisense orientation. To date, CUTs have a relatively uncharacterized function but have been implicated in a number of putative gene regulation and silencing pathways. Thousands of loci leading to the generation of CUTs have been described in the yeast genome. Additionally, stable uncharacterized transcripts, or SUTs, have also been detected in cells and bear many similarities to CUTs but are not degraded through the same pathways.

Chromatin assembly factor-1 (CAF-1) is a protein complex — including Chaf1a (p150), Chaf1b (p60), and p50 subunits — that assembles histone tetramers onto replicating DNA. CAF-1 functions as a histone chaperone that mediates the first step in nucleosome formation by tetramerizing and depositing newly synthesized histone H3/H4 onto DNA rapidly behind replication forks. Several studies have shown that the interaction between CAF-1 and PCNA, which stabilizes CAF-1 at replication forks, is important for CAF-1's role in nucleosome assembly
Danny Reinberg is a Chilean researcher and professor of Biochemistry. His major research focus areas include transcription factors, genetic transcription, histones, RNA Polymerase II, and Chromatin.
References
- ↑ Winkler, Duane D.; Luger, Karolin (2011-05-27). "The Histone Chaperone FACT: Structural Insights and Mechanisms for Nucleosome Reorganization". The Journal of Biological Chemistry. 286 (21): 18369–18374. doi: 10.1074/jbc.R110.180778 . ISSN 0021-9258. PMC 3099653 . PMID 21454601.
- ↑ Martin, Benjamin J. E.; Chruscicki, Adam T.; Howe, LeAnn J. (2018-11-01). "Transcription Promotes the Interaction of the FAcilitates Chromatin Transactions (FACT) Complex with Nucleosomes in Saccharomyces cerevisiae". Genetics. 210 (3): 869–881. doi: 10.1534/genetics.118.301349 . ISSN 0016-6731. PMC 6218215 . PMID 30237209.
- ↑ Frost, Jennifer M.; Kim, M. Yvonne; Park, Guen Tae; Hsieh, Ping-Hung; Nakamura, Miyuki; Lin, Samuel J. H.; Yoo, Hyunjin; Choi, Jaemyung; Ikeda, Yoko; Kinoshita, Tetsu; Choi, Yeonhee (2018-05-15). "FACT complex is required for DNA demethylation at heterochromatin during reproduction in Arabidopsis". Proceedings of the National Academy of Sciences. 115 (20): E4720–E4729. doi: 10.1073/pnas.1713333115 . ISSN 0027-8424. PMC 5960277 . PMID 29712855.
- ↑ Orphanides, George; LeRoy, Gary; Chang, Chun-Hsiang; Luse, Donal S.; Reinberg, Danny (1998). "FACT, a Factor that Facilitates Transcript Elongation through Nucleosomes". Cell. 92 (1): 105–116. doi: 10.1016/S0092-8674(00)80903-4 . PMID 9489704. S2CID 11248520.
- ↑ Orphanides, George; Wu, Wei-Hua; Lane, William S.; Hampsey, Michael; Reinberg, Danny (1999). "The chromatin-specific transcription elongation factor FACT comprises human SPT16 and SSRP1 proteins". Nature. 400 (6741): 284–288. Bibcode:1999Natur.400..284O. doi:10.1038/22350. PMID 10421373. S2CID 4300397.
- ↑ Belotserkovskaya, Rimma; Oh, Sangtaek; Bondarenko, Vladimir A.; Orphanides, George; Studitsky, Vasily; Reinberg, Danny (2003). "FACT facilitates transcription-dependent nucleosome alteration". Science. 301 (5636): 1090–1093. Bibcode:2003Sci...301.1090B. doi:10.1126/science.1085703. PMID 12934006. S2CID 26667338.
- ↑ Saunders, Abbie; Werner, Janis; Andrulis, Erik D.; Nakayama, Takahiro; Hirose, Susumu; Reinberg, Danny; Lis, John T. (2003). "Tracking FACT and the RNA Polymerase II Elongation Complex Through Chromatin in Vivo". Science. 301 (5636): 1094–1096. Bibcode:2003Sci...301.1094S. doi:10.1126/science.1085712. PMID 12934007. S2CID 10064786.