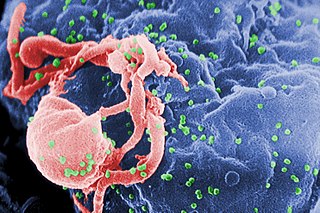
The human immunodeficiency viruses (HIV) are two species of Lentivirus that infect humans. Over time, they cause acquired immunodeficiency syndrome (AIDS), a condition in which progressive failure of the immune system allows life-threatening opportunistic infections and cancers to thrive. Without treatment, average survival time after infection with HIV is estimated to be 9 to 11 years, depending on the HIV subtype. In most cases, HIV is a sexually transmitted infection and occurs by contact with or transfer of blood, pre-ejaculate, semen, and vaginal fluids. Research has shown that HIV is untransmittable through condomless sexual intercourse if the HIV-positive partner has a consistently undetectable viral load. Non-sexual transmission can occur from an infected mother to her infant during pregnancy, during childbirth by exposure to her blood or vaginal fluid, and through breast milk. Within these bodily fluids, HIV is present as both free virus particles and virus within infected immune cells.
A provirus is a virus genome that is integrated into the DNA of a host cell. In the case of bacterial viruses (bacteriophages), proviruses are often referred to as prophages. However, it is important to note that proviruses are distinctly different from prophages and these terms should not be used interchangeably. Unlike prophages, proviruses do not excise themselves from the host genome when the host cell is stressed.

A retrovirus is a type of virus that inserts a copy of its RNA genome into the DNA of a host cell that it invades, thus changing the genome of that cell. Once inside the host cell's cytoplasm, the virus uses its own reverse transcriptase enzyme to produce DNA from its RNA genome, the reverse of the usual pattern, thus retro (backwards). The new DNA is then incorporated into the host cell genome by an integrase enzyme, at which point the retroviral DNA is referred to as a provirus. The host cell then treats the viral DNA as part of its own genome, transcribing and translating the viral genes along with the cell's own genes, producing the proteins required to assemble new copies of the virus.
Antiviral drugs are a class of medication used for treating viral infections. Most antivirals target specific viruses, while a broad-spectrum antiviral is effective against a wide range of viruses. Unlike most antibiotics, antiviral drugs do not destroy their target pathogen; instead they inhibit its development.
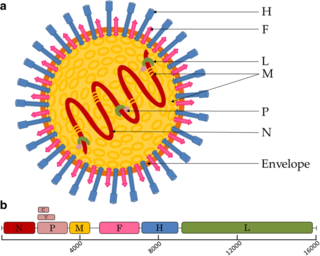
Paramyxoviridae is a family of negative-strand RNA viruses in the order Mononegavirales. Vertebrates serve as natural hosts. Diseases associated with this family include measles, mumps, and respiratory tract infections. The family has four subfamilies, 17 genera, and 77 species, three genera of which are unassigned to a subfamily.

Coronaviruses are a group of related RNA viruses that cause diseases in mammals and birds. In humans and birds, they cause respiratory tract infections that can range from mild to lethal. Mild illnesses in humans include some cases of the common cold, while more lethal varieties can cause SARS, MERS, and COVID-19. In cows and pigs they cause diarrhea, while in mice they cause hepatitis and encephalomyelitis.

Severe acute respiratory syndrome–related coronavirus is a species of coronavirus that infects humans, bats and certain other mammals. It is an enveloped positive-sense single-stranded RNA virus that enters its host cell by binding to the angiotensin-converting enzyme 2 (ACE2) receptor. It is a member of the genus Betacoronavirus and subgenus Sarbecovirus.

Reoviridae is a family of double-stranded RNA viruses. Member viruses have a wide host range, including vertebrates, invertebrates, plants, protists and fungi. They lack lipid envelopes and package their segmented genome within multi-layered capsids. Lack of a lipid envelope has allowed three-dimensional structures of these large complex viruses to be obtained, revealing a structural and likely evolutionary relationship to the cystovirus family of bacteriophage. There are currently 97 species in this family, divided among 15 genera in two subfamilies. Reoviruses can affect the gastrointestinal system and respiratory tract. The name "reo-" is an acronym for "respiratory enteric orphan" viruses. The term "orphan virus" refers to the fact that some of these viruses have been observed not associated with any known disease. Even though viruses in the family Reoviridae have more recently been identified with various diseases, the original name is still used.

Adenoviruses are medium-sized, nonenveloped viruses with an icosahedral nucleocapsid containing a double stranded DNA genome. Their name derives from their initial isolation from human adenoids in 1953.
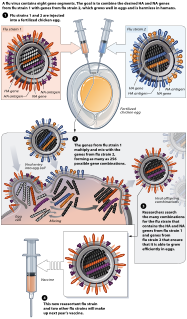
Reassortment is the mixing of the genetic material of a species into new combinations in different individuals. Several different processes contribute to reassortment, including assortment of chromosomes, and chromosomal crossover. It is particularly used when two similar viruses that are infecting the same cell exchange genetic material. In particular, reassortment occurs among influenza viruses, whose genomes consist of eight distinct segments of RNA. These segments act like mini-chromosomes, and each time a flu virus is assembled, it requires one copy of each segment.

Geminiviridae is a family of plant viruses that encode their genetic information on a circular genome of single-stranded (ss) DNA. There are currently 485 species in this family, divided among 9 genera. Diseases associated with this family include: bright yellow mosaic, yellow mosaic, yellow mottle, leaf curling, stunting, streaks, reduced yields. They have single-stranded circular DNA genomes encoding genes that diverge in both directions from a virion strand origin of replication. According to the Baltimore classification they are considered class II viruses. It is the largest known family of single stranded DNA viruses.
Viral pathogenesis is the study of the process and mechanisms by which viruses cause diseases in their target hosts, often at the cellular or molecular level. It is a specialized field of study in virology.

Viral replication is the formation of biological viruses during the infection process in the target host cells. Viruses must first get into the cell before viral replication can occur. Through the generation of abundant copies of its genome and packaging these copies, the virus continues infecting new hosts. Replication between viruses is greatly varied and depends on the type of genes involved in them. Most DNA viruses assemble in the nucleus while most RNA viruses develop solely in cytoplasm.

Herpesviridae is a large family of DNA viruses that cause infections and certain diseases in animals, including humans. The members of this family are also known as herpesviruses. The family name is derived from the Greek word herpein 'to creep', referring to spreading cutaneous lesions, usually involving blisters, seen in flares of herpes simplex 1, herpes simplex 2 and herpes zoster (shingles). In 1971, the International Committee on the Taxonomy of Viruses (ICTV) established Herpesvirus as a genus with 23 viruses among four groups. As of 2020, 107 species are recognized, all but one of which are in one of the three subfamilies. Herpesviruses can cause both latent or lytic infections.
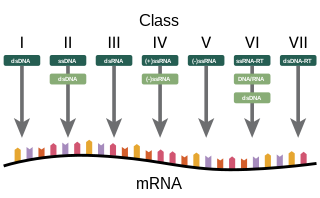
Baltimore classification is a system used to classify viruses based on their manner of messenger RNA (mRNA) synthesis. By organizing viruses based on their manner of mRNA production, it is possible to study viruses that behave similarly as a distinct group. Seven Baltimore groups are described that take into consideration whether the viral genome is made of deoxyribonucleic acid (DNA) or ribonucleic acid (RNA), whether the genome is single- or double-stranded, and whether the sense of a single-stranded RNA genome is positive or negative.
Viral vectors are tools commonly used by molecular biologists to deliver genetic material into cells. This process can be performed inside a living organism or in cell culture. Viruses have evolved specialized molecular mechanisms to efficiently transport their genomes inside the cells they infect. Delivery of genes or other genetic materia by a vector is termed transduction and the infected cells are described as transduced. Molecular biologists first harnessed this machinery in the 1970s. Paul Berg used a modified SV40 virus containing DNA from the bacteriophage λ to infect monkey kidney cells maintained in culture.

A virus is a tiny infectious agent that reproduces inside the cells of living hosts. When infected, the host cell is forced to rapidly produce thousands of identical copies of the original virus. Unlike most living things, viruses do not have cells that divide; new viruses assemble in the infected host cell. But unlike simpler infectious agents like prions, they contain genes, which allow them to mutate and evolve. Over 4,800 species of viruses have been described in detail out of the millions in the environment. Their origin is unclear: some may have evolved from plasmids—pieces of DNA that can move between cells—while others may have evolved from bacteria.

A virus is a submicroscopic infectious agent that replicates only inside the living cells of an organism. Viruses infect all types of life forms, from animals and plants to microorganisms, including bacteria and archaea. Since Dmitri Ivanovsky's 1892 article describing a non-bacterial pathogen infecting tobacco plants and the discovery of the tobacco mosaic virus by Martinus Beijerinck in 1898, more than 6,000 virus species have been described in detail of the millions of types of viruses in the environment. Viruses are found in almost every ecosystem on Earth and are the most numerous type of biological entity. The study of viruses is known as virology, a subspeciality of microbiology.
In biology, a pathogen in the oldest and broadest sense, is any organism that can produce disease. A pathogen may also be referred to as an infectious agent, or simply a germ.
Monodnaviria is a realm of viruses that includes all single-stranded DNA viruses that encode an endonuclease of the HUH superfamily that initiates rolling circle replication of the circular viral genome. Viruses descended from such viruses are also included in the realm, including certain linear single-stranded DNA (ssDNA) viruses and circular double-stranded DNA (dsDNA) viruses. These atypical members typically replicate through means other than rolling circle replication.












