
Mimivirus is a genus of giant viruses, in the family Mimiviridae. Amoeba serve as their natural hosts. This genus contains a single identified species named Acanthamoeba polyphaga mimivirus (APMV). It also refers to a group of phylogenetically related large viruses.

The mobilome is the entire set of mobile genetic elements in a genome. Mobilomes are found in eukaryotes, prokaryotes, and viruses. The compositions of mobilomes differ among lineages of life, with transposable elements being the major mobile elements in eukaryotes, and plasmids and prophages being the major types in prokaryotes. Virophages contribute to the viral mobilome.

Asfarviridae is a family of viruses, the best-studied of which is African swine fever virus. They are double-stranded DNA viruses.

Virophages are small, double-stranded DNA viral phages that require the co-infection of another virus. The co-infecting viruses are typically giant viruses. Virophages rely on the viral replication factory of the co-infecting giant virus for their own replication. One of the characteristics of virophages is that they have a parasitic relationship with the co-infecting virus. Their dependence upon the giant virus for replication often results in the deactivation of the giant viruses. The virophage may improve the recovery and survival of the host organism.

Mimivirus-dependent virus Sputnik is a subviral agent that reproduces in amoeba cells that are already infected by a certain helper virus; Sputnik uses the helper virus's machinery for reproduction and inhibits replication of the helper virus. It is known as a virophage, in analogy to the term bacteriophage.

Mimiviridae is a family of viruses. Amoeba and other protists serve as natural hosts. The family is divided in up to 4 subfamilies. Viruses in this family belong to the nucleocytoplasmic large DNA virus clade (NCLDV), also referred to as giant viruses.
Mamavirus is a large and complex virus in the Group I family Mimiviridae. The virus is exceptionally large, and larger than many bacteria. Mamavirus and other mimiviridae belong to nucleocytoplasmic large DNA virus (NCLDVs) family. Mamavirus can be compared to the similar complex virus mimivirus; mamavirus was so named because it is similar to but larger than mimivirus.
Marseillevirus is a genus of viruses, in the family Marseilleviridae. There are two species in this genus. It is the prototype of a family of nucleocytoplasmic large DNA viruses (NCLDV) of eukaryotes. It was isolated from amoeba.

Cafeteria roenbergensis virus (CroV) is a giant virus that infects the marine bicosoecid flagellate Cafeteria roenbergensis, a member of the microzooplankton community.
A giant virus, sometimes referred to as a girus, is a very large virus, some of which are larger than typical bacteria. All known giant viruses belong to the phylum Nucleocytoviricota.

Marseilleviridae is a family of viruses first named in 2012. The genomes of these viruses are double-stranded DNA. Amoeba are often hosts, but there is evidence that they are found in humans as well. The family contains one genus and four species, two of which are unassigned to a genus. It is a member of the nucleocytoplasmic large DNA viruses clade.

Megavirus is a viral genus containing a single identified species named Megavirus chilense, phylogenetically related to Acanthamoeba polyphaga Mimivirus (APMV). In colloquial speech, Megavirus chilensis is more commonly referred to as just “Megavirus”. Until the discovery of pandoraviruses in 2013, it had the largest capsid diameter of all known viruses, as well as the largest and most complex genome among all known viruses.

Pandoravirus is a genus of giant virus, first discovered in 2013. It is the second largest in physical size of any known viral genus. Pandoraviruses have double stranded DNA genomes, with the largest genome size of any known viral genus.
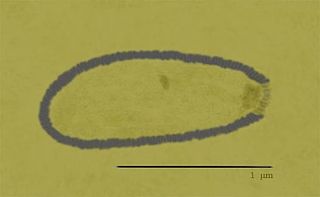
Pithovirus, first described in a 2014 paper, is a genus of giant virus known from two species, Pithovirus sibericum, which infects amoebas and Pithovirus massiliensis. It is a double-stranded DNA virus and is a member of the nucleocytoplasmic large DNA viruses clade. The 2014 discovery was made when a viable specimen was found in a 30,000-year-old ice core harvested from permafrost in Siberia, Russia.
Chlorovirus, also known as Chlorella virus, is a genus of giant double-stranded DNA viruses, in the family Phycodnaviridae. This genus is found globally in freshwater environments where freshwater microscopic algae serve as natural hosts. There are 19 species in this genus.

Mimivirus-dependent virus Zamilon, or Zamilon, is a virophage, a group of small DNA viruses that infect protists and require a helper virus to replicate; they are a type of satellite virus. Discovered in 2013 in Tunisia, infecting Acanthamoeba polyphaga amoebae, Zamilon most closely resembles Sputnik, the first virophage to be discovered. The name is Arabic for "the neighbour". Its spherical particle is 50–60 nm in diameter, and contains a circular double-stranded DNA genome of around 17 kb, which is predicted to encode 20 polypeptides. A related strain, Zamilon 2, has been identified in North America.

Tupanvirus is a genus of viruses first described in 2018. The genus is composed of two species of virus that are in the giant virus group. Researchers discovered the first isolate in 2012 from deep water sediment samples taken at 3000m depth off the coast of Brazil. The second isolate was collected from a soda lake in Southern Nhecolândia, Brazil in 2014. They are named after Tupã (Tupan), a Guaraní thunder god, and the places they were found. These are the first viruses reported to possess genes for amino-acyl tRNA synthetases for all 20 standard amino acids.

Medusavirus is a nucleocytoplasmic large DNA virus first isolated from a Japanese hot spring in 2019. It notably encodes all five types of histones — H1, H2A, H2B, H3, and H4 — which are involved in DNA packaging in eukaryotes, raising the possibility that they may have been involved in the origin of eukaryotes. The virus can harden amoebas of the species Acanthamoeba castellanii into stone-like cysts, but infection usually causes infected amoebas to burst open. The virus was named after Medusa, the monster in Greek mythology whose gaze turned people to stone.

Varidnaviria is a realm of viruses that includes all DNA viruses that encode major capsid proteins that contain a vertical jelly roll fold. The major capsid proteins (MCP) form into pseudohexameric subunits of the viral capsid, which stores the viral deoxyribonucleic acid (DNA), and are perpendicular, or vertical, to the surface of the capsid. Apart from this, viruses in the realm also share many other characteristics, such as minor capsid proteins (mCP) with the vertical jelly roll fold, an ATPase that packages viral DNA into the capsid, and a DNA polymerase that replicates the viral genome.
Nucleocytoviricota is a phylum of viruses. Members of the phylum are also known as the nucleocytoplasmic large DNA viruses (NCLDV), which serves as the basis of the name of the phylum with the suffix -viricota for virus phylum. These viruses are referred to as nucleocytoplasmic because they are often able to replicate in both the host's cell nucleus and cytoplasm.














