Design
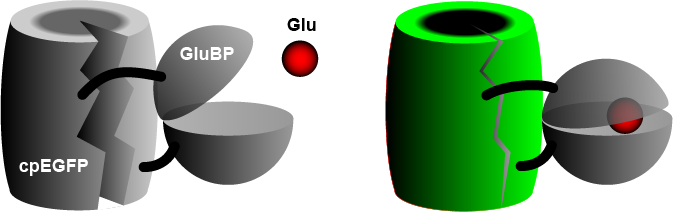
The widely used iGluSnFR consists of a circularly permuted enhanced green fluorescent protein (cpEGFP) fused to a glutamate binding protein (GluBP) from a bacterium. [3] When GluBP binds a glutamate molecule, it changes its shape, pulling the EGFP barrel together, increasing fluorescence. A specific peptide segment (PDGFR) is included to bring the sensor to the outside of the cell membrane. [4] In the more recent version by Aggarwal et al. (2022), [1] researchers introduced iGluSnFR to two additional anchoring domains, a glycosylphostidylinositol (GPI) anchor, and a modified form of the cytosolic -cterminal domain of Stargazin with a PDZ ligand.