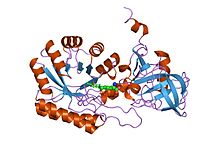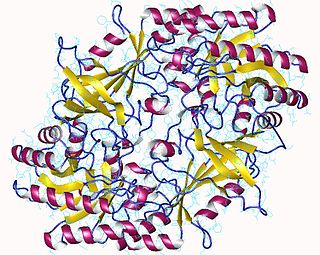
The enzyme ornithine decarboxylase (ODC) catalyzes the decarboxylation of ornithine to form putrescine. This reaction is the committed step in polyamine synthesis. In humans, this protein has 461 amino acids and forms a homodimer.

Pyridoxal phosphate (PLP, pyridoxal 5'-phosphate, P5P), the active form of vitamin B6, is a coenzyme in a variety of enzymatic reactions. The Enzyme commission has catalogued more than 140 PLP-dependent activities, corresponding to ~4% of all classified activities. The versatility of PLP arises from its ability to covalently bind the substrate, and then to act as an electrophilic catalyst, thereby stabilizing different types of carbanionic reaction intermediates.
Biosynthesis is a multi-step, enzyme-catalyzed process where substrates are converted into more complex products in living organisms. In biosynthesis, simple compounds are modified, converted into other compounds, or joined together to form macromolecules. This process often consists of metabolic pathways. Some of these biosynthetic pathways are located within a single cellular organelle, while others involve enzymes that are located within multiple cellular organelles. Examples of these biosynthetic pathways include the production of lipid membrane components and nucleotides. Biosynthesis is usually synonymous with anabolism.

Amino acid synthesis is the set of biochemical processes by which the amino acids are produced. The substrates for these processes are various compounds in the organism's diet or growth media. Not all organisms are able to synthesize all amino acids. For example, humans can only synthesize 11 of the 20 standard amino acids, and in time of accelerated growth, histidine can be considered an essential amino acid.

Carbamoyl phosphate synthetase catalyzes the ATP-dependent synthesis of carbamoyl phosphate from glutamine or ammonia and bicarbonate. This enzyme catalyzes the reaction of ATP and bicarbonate to produce carboxy phosphate and ADP. Carboxy phosphate reacts with ammonia to give carbamic acid. In turn, carbamic acid reacts with a second ATP to give carbamoyl phosphate plus ADP.
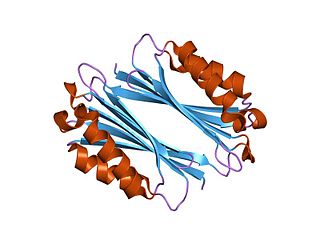
Adenosylmethionine decarboxylase is an enzyme that catalyzes the conversion of S-adenosyl methionine to S-adenosylmethioninamine. Polyamines such as spermidine and spermine are essential for cellular growth under most conditions, being implicated in many cellular processes including DNA, RNA and protein synthesis. S-adenosylmethionine decarboxylase (AdoMetDC) plays an essential regulatory role in the polyamine biosynthetic pathway by generating the n-propylamine residue required for the synthesis of spermidine and spermine from putrescein. Unlike many amino acid decarboxylases AdoMetDC uses a covalently bound pyruvate residue as a cofactor rather than the more common pyridoxal 5'-phosphate. These proteins can be divided into two main groups which show little sequence similarity either to each other, or to other pyruvoyl-dependent amino acid decarboxylases: class I enzymes found in bacteria and archaea, and class II enzymes found in eukaryotes. In both groups the active enzyme is generated by the post-translational autocatalytic cleavage of a precursor protein. This cleavage generates the pyruvate precursor from an internal serine residue and results in the formation of two non-identical subunits termed alpha and beta which form the active enzyme.
In enzymology, a 2,4-diaminopentanoate dehydrogenase (EC 1.4.1.12) is an enzyme that catalyzes the chemical reaction

In enzymology, an alanine racemase is an enzyme that catalyzes the chemical reaction
In enzymology, a diaminopimelate epimerase is an enzyme that catalyzes the chemical reaction

Cystathionine beta-lyase, also commonly referred to as CBL or β-cystathionase, is an enzyme that primarily catalyzes the following α,β-elimination reaction

Acid-Induced Arginine Decarboxylase (AdiA), also commonly referred to as arginine decarboxylase, is an enzyme responsible for catalyzing the conversion of L-arginine into agmatine and carbon dioxide. The process consumes a proton in the decarboxylation and employs a pyridoxal-5'-phosphate (PLP) cofactor, similar to other enzymes involved in amino acid metabolism, such as ornithine decarboxylase and glutamine decarboxylase. It is found in bacteria and virus, though most research has so far focused on forms of the enzyme in bacteria. During the AdiA catalyzed decarboxylation of arginine, the necessary proton is consumed from the cell cytoplasm which helps to prevent the over-accumulation of protons inside the cell and serves to increase the intracellular pH. Arginine decarboxylase is part of an enzymatic system in Escherichia coli, Salmonella Typhimurium, and methane-producing bacteria Methanococcus jannaschii that makes these organisms acid resistant and allows them to survive under highly acidic medium.

In enzymology, diaminopimelate decarboxylase, also known as diaminopimelic acid decarboxylase, DAPDC, meso-diaminopimelate decarboxylase, DAP-decarboxylase, and meso-2,6-diaminoheptanedioate carboxy-lyase, is an enzyme that catalyzes the cleavage of carbon-carbon bonds in meso 2,6 diaminoheptanedioate to produce CO2 and L-lysine, the essential amino acid. It employs the cofactor pyridoxal phosphate, also known as PLP, which participates in numerous enzymatic transamination, decarboxylation and deamination reactions.
The Walker A and Walker B motifs are protein sequence motifs, known to have highly conserved three-dimensional structures. These were first reported in ATP-binding proteins by Walker and co-workers in 1982.
Aminotransferase class-V is an evolutionary conserved protein domain. This domain is found in amino transferases, and other enzymes including cysteine desulphurase EC:4.4.1.-.
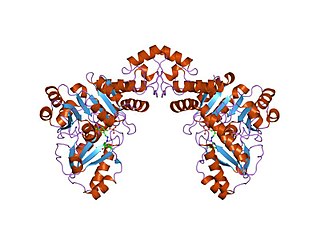
In molecular biology, the Cys/Met metabolism PLP-dependent enzyme family is a family of proteins including enzymes involved in cysteine and methionine metabolism which use PLP (pyridoxal-5'-phosphate) as a cofactor.

In molecular biology, group III pyridoxal-dependent decarboxylases are a family of bacterial enzymes comprising ornithine decarboxylase EC 4.1.1.17, lysine decarboxylase EC 4.1.1.18 and arginine decarboxylase EC 4.1.1.19.
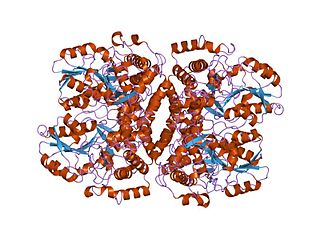
In molecular biology, the group I pyridoxal-dependent decarboxylases, also known as glycine cleavage system P-proteins, are a family of enzymes consisting of glycine cleavage system P-proteins EC 1.4.4.2 from bacterial, mammalian and plant sources. The P protein is part of the glycine decarboxylase multienzyme complex (GDC) also annotated as glycine cleavage system or glycine synthase. The P protein binds the alpha-amino group of glycine through its pyridoxal phosphate cofactor, carbon dioxide is released and the remaining methylamin moiety is then transferred to the lipoamide cofactor of the H protein. GDC consists of four proteins P, H, L and T.
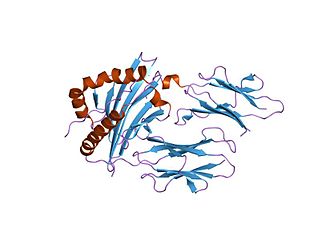
In molecular biology, group II pyridoxal-dependent decarboxylases are family of enzymes including aromatic-L-amino-acid decarboxylase EC 4.1.1.28, which catalyses the decarboxylation of tryptophan to tryptamine, tyrosine decarboxylase EC 4.1.1.25, which converts tyrosine into tyramine and histidine decarboxylase EC 4.1.1.22, which catalyses the decarboxylation of histidine to histamine.
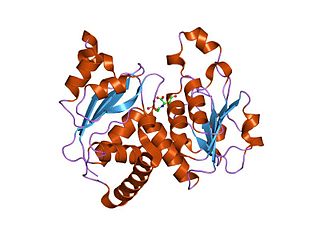
In molecular biology, the ATCase/OTCase family is a protein family which contains two related enzymes: aspartate carbamoyltransferase EC 2.1.3.2 and ornithine carbamoyltransferase EC 2.1.3.3. It has been shown that these enzymes are evolutionary related. The predicted secondary structure of both enzymes is similar and there are some regions of sequence similarities. One of these regions includes three residues which have been shown, by crystallographic studies to be implicated in binding the phosphoryl group of carbamoyl phosphate and may also play a role in trimerisation of the molecules. The N-terminal domain is the carbamoyl phosphate binding domain. The C-terminal domain is an aspartate/ornithine-binding domain.
Glutamate 2,3-aminomutase is an enzyme that belongs to the radical s-adenosyl methionine (SAM) superfamily. Radical SAM enzymes facilitate the reductive cleavage of S-adenosylmethionine (SAM) through the use of radical chemistry and an iron-sulfur cluster. This enzyme family is implicated in the biosynthesis of DNA precursors, vitamin, cofactor, antibiotic and herbicides and in biodegradation pathways. In particular, glutamate 2,3 aminomutase is involved in the conversion of L-alpha-glutamate to L-beta-glutamate in Clostridium difficile. The generalized reaction is shown below: