Methionine is an essential amino acid in humans.

Lysine is an α-amino acid that is a precursor to many proteins. It contains an α-amino group, an α-carboxylic acid group, and a side chain lysyl, classifying it as a basic, charged, aliphatic amino acid. It is encoded by the codons AAA and AAG. Like almost all other amino acids, the α-carbon is chiral and lysine may refer to either enantiomer or a racemic mixture of both. For the purpose of this article, lysine will refer to the biologically active enantiomer L-lysine, where the α-carbon is in the S configuration.
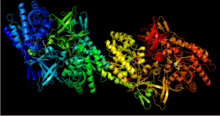
The enzyme ornithine decarboxylase catalyzes the decarboxylation of ornithine to form putrescine. This reaction is the committed step in polyamine synthesis. In humans, this protein has 461 amino acids and forms a homodimer.
Decarboxylation is a chemical reaction that removes a carboxyl group and releases carbon dioxide (CO2). Usually, decarboxylation refers to a reaction of carboxylic acids, removing a carbon atom from a carbon chain. The reverse process, which is the first chemical step in photosynthesis, is called carboxylation, the addition of CO2 to a compound. Enzymes that catalyze decarboxylations are called decarboxylases or, the more formal term, carboxy-lyases (EC number 4.1.1).

Aromatic L-amino acid decarboxylase, also known as DOPA decarboxylase (DDC), tryptophan decarboxylase, and 5-hydroxytryptophan decarboxylase, is a lyase enzyme, located in region 7p12.2-p12.1.

Pyridoxal phosphate (PLP, pyridoxal 5'-phosphate, P5P), the active form of vitamin B6, is a coenzyme in a variety of enzymatic reactions. The International Union of Biochemistry and Molecular Biology has catalogued more than 140 PLP-dependent activities, corresponding to ~4% of all classified activities. The versatility of PLP arises from its ability to covalently bind the substrate, and then to act as an electrophilic catalyst, thereby stabilizing different types of carbanionic reaction intermediates.
Biosynthesis, i.e., chemical synthesis occurring in biological contexts, is a term most often referring to multi-step, enzyme-catalyzed processes where chemical substances absorbed as nutrients serve as enzyme substrates, with conversion by the living organism either into simpler or more complex products. Examples of biosynthetic pathways include those for the production of amino acids, lipid membrane components, and nucleotides, but also for the production of all classes of biological macromolecules, and of acetyl-coenzyme A, adenosine triphosphate, nicotinamide adenine dinucleotide and other key intermediate and transactional molecules needed for metabolism. Thus, in biosynthesis, any of an array of compounds, from simple to complex, are converted into other compounds, and so it includes both the catabolism and anabolism of complex molecules. Biosynthetic processes are often represented via charts of metabolic pathways. A particular biosynthetic pathway may be located within a single cellular organelle, while others involve enzymes that are located across an array of cellular organelles and structures.
Carboxy-lyases, also known as decarboxylases, are carbon–carbon lyases that add or remove a carboxyl group from organic compounds. These enzymes catalyze the decarboxylation of amino acids, beta-keto acids and alpha-keto acids.

In enzymology, an alanine racemase is an enzyme that catalyzes the chemical reaction
In enzymology, an aspartate racemase is an enzyme that catalyzes the following chemical reaction:
In enzymology, a diaminopimelate epimerase is an enzyme that catalyzes the chemical reaction

Cystathionine beta-lyase, also commonly referred to as CBL or β-cystathionase, is an enzyme that primarily catalyzes the following α,β-elimination reaction

Threonine ammonia-lyase (EC 4.3.1.19, systematic name L-threonine ammonia-lyase (2-oxobutanoate-forming), also commonly referred to as threonine deaminase or threonine dehydratase, is an enzyme responsible for catalyzing the conversion of L-threonine into α-ketobutyrate and ammonia:

The enzyme Acid-Induced Arginine Decarboxylase (AdiA), also commonly referred to as arginine decarboxylase, catalyzes the conversion of L-arginine into agmatine and carbon dioxide. The process consumes a proton in the decarboxylation and employs a pyridoxal-5'-phosphate (PLP) cofactor, similar to other enzymes involved in amino acid metabolism, such as ornithine decarboxylase and glutamine decarboxylase. It is found in bacteria and virus, though most research has so far focused on forms of the enzyme in bacteria. During the AdiA catalyzed decarboxylation of arginine, the necessary proton is consumed from the cell cytoplasm which helps to prevent the over-accumulation of protons inside the cell and serves to increase the intracellular pH. Arginine decarboxylase is part of an enzymatic system in Escherichia coli, Salmonella Typhimurium, and methane-producing bacteria Methanococcus jannaschii that makes these organisms acid resistant and allows them to survive under highly acidic medium.

Diphosphomevalonate decarboxylase (EC 4.1.1.33), most commonly referred to in scientific literature as mevalonate diphosphate decarboxylase, is an enzyme that catalyzes the chemical reaction

In enzymology, a serine C-palmitoyltransferase (EC 2.3.1.50) is an enzyme that catalyzes the chemical reaction:

Diaminopimelic acid (DAP) is an amino acid, representing an epsilon-carboxy derivative of lysine. meso-α,ε-Diaminopimelic acid is the last intermediate in the biosynthesis of lysine and undergoes decarboxylation by diaminopimelate decarboxylase to give the final product.
In molecular biology, the Cys/Met metabolism PLP-dependent enzyme family is a family of proteins including enzymes involved in cysteine and methionine metabolism which use PLP (pyridoxal-5'-phosphate) as a cofactor.

In molecular biology, group III pyridoxal-dependent decarboxylases are a family of bacterial enzymes comprising ornithine decarboxylase EC 4.1.1.17, lysine decarboxylase EC 4.1.1.18 and arginine decarboxylase EC 4.1.1.19.

In molecular biology, group IV pyridoxal-dependent decarboxylases are a family of enzymes comprising ornithine decarboxylase EC 4.1.1.17, lysine decarboxylase EC 4.1.1.18, arginine decarboxylase EC 4.1.1.19 and diaminopimelate decarboxylaseEC 4.1.1.20. It is also known as the Orn/Lys/Arg decarboxylase class-II family.