A retrovirus is a type of virus that inserts a DNA copy of its RNA genome into the DNA of a host cell that it invades, thus changing the genome of that cell. After invading a host cell's cytoplasm, the virus uses its own reverse transcriptase enzyme to produce DNA from its RNA genome, the reverse of the usual pattern, thus retro (backwards). The new DNA is then incorporated into the host cell genome by an integrase enzyme, at which point the retroviral DNA is referred to as a provirus. The host cell then treats the viral DNA as part of its own genome, transcribing and translating the viral genes along with the cell's own genes, producing the proteins required to assemble new copies of the virus. Many retroviruses cause serious diseases in humans, other mammals, and birds.

Totiviridae is a family of double-stranded RNA viruses. Giardia lamblia, leishmania, trichomonas vaginalis, and fungi serve as natural hosts. The name of the group derives from Latin toti which means undivided or whole. There are 28 species in this family, assigned to 5 genera.

Barnaviridae is a family of non-enveloped, positive-strand RNA viruses. Cultivated mushrooms serve as natural hosts. The family has one genus, Barnavirus, which contains one species: Mushroom bacilliform virus. Diseases associated with this family includes La France disease.

Viral replication is the formation of biological viruses during the infection process in the target host cells. Viruses must first get into the cell before viral replication can occur. Through the generation of abundant copies of its genome and packaging these copies, the virus continues infecting new hosts. Replication between viruses is greatly varied and depends on the type of genes involved in them. Most DNA viruses assemble in the nucleus while most RNA viruses develop solely in cytoplasm.
The genome and proteins of HIV (human immunodeficiency virus) have been the subject of extensive research since the discovery of the virus in 1983. "In the search for the causative agent, it was initially believed that the virus was a form of the Human T-cell leukemia virus (HTLV), which was known at the time to affect the human immune system and cause certain leukemias. However, researchers at the Pasteur Institute in Paris isolated a previously unknown and genetically distinct retrovirus in patients with AIDS which was later named HIV." Each virion comprises a viral envelope and associated matrix enclosing a capsid, which itself encloses two copies of the single-stranded RNA genome and several enzymes. The discovery of the virus itself occurred two years following the report of the first major cases of AIDS-associated illnesses.
Ribosome shunting is a mechanism of translation initiation in which ribosomes bypass, or "shunt over", parts of the 5' untranslated region to reach the start codon. However, a benefit of ribosomal shunting is that it can translate backwards allowing more information to be stored than usual in an mRNA molecule. Some viral RNAs have been shown to use ribosome shunting as a more efficient form of translation during certain stages of viral life cycle or when translation initiation factors are scarce. Some viruses known to use this mechanism include adenovirus, Sendai virus, human papillomavirus, duck hepatitis B pararetrovirus, rice tungro bacilliform viruses, and cauliflower mosaic virus. In these viruses the ribosome is directly translocated from the upstream initiation complex to the start codon (AUG) without the need to unwind RNA secondary structures.
Eukaryotic translation is the biological process by which messenger RNA is translated into proteins in eukaryotes. It consists of four phases: initiation, elongation, termination, and recapping.
In genetics, attenuation is a regulatory mechanism for some bacterial operons that results in premature termination of transcription. The canonical example of attenuation used in many introductory genetics textbooks, is ribosome-mediated attenuation of the trp operon. Ribosome-mediated attenuation of the trp operon relies on the fact that, in bacteria, transcription and translation proceed simultaneously. Attenuation involves a provisional stop signal (attenuator), located in the DNA segment that corresponds to the leader sequence of mRNA. During attenuation, the ribosome becomes stalled (delayed) in the attenuator region in the mRNA leader. Depending on the metabolic conditions, the attenuator either stops transcription at that point or allows read-through to the structural gene part of the mRNA and synthesis of the appropriate protein.
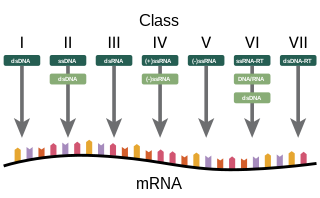
Baltimore classification is a system used to classify viruses based on their manner of messenger RNA (mRNA) synthesis. By organizing viruses based on their manner of mRNA production, it is possible to study viruses that behave similarly as a distinct group. Seven Baltimore groups are described that take into consideration whether the viral genome is made of deoxyribonucleic acid (DNA) or ribonucleic acid (RNA), whether the genome is single- or double-stranded, and whether the sense of a single-stranded RNA genome is positive or negative.
Group-specific antigen, or gag, is the polyprotein that contains the core structural proteins of an Ortervirus. It was named as such because scientists used to believe it was antigenic. Now it is known that it makes up the inner shell, not the envelope exposed outside. It makes up all the structural units of viral conformation and provides supportive framework for mature virion.

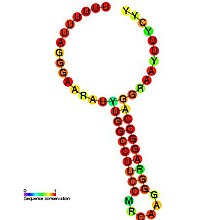
In molecular biology, the coronavirus frameshifting stimulation element is a conserved stem-loop of RNA found in coronaviruses that can promote ribosomal frameshifting. Such RNA molecules interact with a downstream region to form a pseudoknot structure; the region varies according to the virus but pseudoknot formation is known to stimulate frameshifting. In the classical situation, a sequence 32 nucleotides downstream of the stem is complementary to part of the loop. In other coronaviruses, however, another stem-loop structure around 150 nucleotides downstream can interact with members of this family to form kissing stem-loops and stimulate frameshifting.

The retroviral psi packaging element, also known as the Ψ RNA packaging signal, is a cis-acting RNA element identified in the genomes of the retroviruses Human immunodeficiency virus (HIV) and Simian immunodeficiency virus (SIV). It is involved in regulating the essential process of packaging the retroviral RNA genome into the viral capsid during replication. The final virion contains a dimer of two identical unspliced copies of the viral genome.
Ribosomal frameshifting, also known as translational frameshifting or translational recoding, is a biological phenomenon that occurs during translation that results in the production of multiple, unique proteins from a single mRNA. The process can be programmed by the nucleotide sequence of the mRNA and is sometimes affected by the secondary, 3-dimensional mRNA structure. It has been described mainly in viruses, retrotransposons and bacterial insertion elements, and also in some cellular genes.

Sobemovirus is a genus of viruses. Plants serve as natural hosts. There are 20 species in this genus. Diseases associated with this genus include: mosaics and mottles.

Rev is a transactivating protein that is essential to the regulation of HIV-1 protein expression. A nuclear localization signal is encoded in the rev gene, which allows the Rev protein to be localized to the nucleus, where it is involved in the export of unspliced and incompletely spliced mRNAs. In the absence of Rev, mRNAs of the HIV-1 late (structural) genes are retained in the nucleus, preventing their translation.
Bovine immunodeficiency virus (BIV) is a retrovirus belonging to the genus Lentivirus. It is similar to the human immunodeficiency virus (HIV) and infects cattle. The cells primarily infected are lymphocytes and monocytes/macrophages.

Ribosomal pause refers to the queueing or stacking of ribosomes during translation of the nucleotide sequence of mRNA transcripts. These transcripts are decoded and converted into an amino acid sequence during protein synthesis by ribosomes. Due to the pause sites of some mRNA's, there is a disturbance caused in translation. Ribosomal pausing occurs in both eukaryotes and prokaryotes. A more severe pause is known as a ribosomal stall.

A slippery sequence is a small section of codon nucleotide sequences that controls the rate and chance of ribosomal frameshifting. A slippery sequence causes a faster ribosomal transfer which in turn can cause the reading ribosome to "slip." This allows a tRNA to shift by 1 base (−1) after it has paired with its anticodon, changing the reading frame. A −1 frameshift triggered by such a sequence is a Programmed −1 Ribosomal Frameshift. It is followed by a spacer region, and an RNA secondary structure. Such sequences are common in virus polyproteins.
ORF1ab refers collectively to two open reading frames (ORFs), ORF1a and ORF1b, that are conserved in the genomes of nidoviruses, a group of viruses that includes coronaviruses. The genes express large polyproteins that undergo proteolysis to form several nonstructural proteins with various functions in the viral life cycle, including proteases and the components of the replicase-transcriptase complex (RTC). Together the two ORFs are sometimes referred to as the replicase gene. They are related by a programmed ribosomal frameshift that allows the ribosome to continue translating past the stop codon at the end of ORF1a, in a -1 reading frame. The resulting polyproteins are known as pp1a and pp1ab.