Structure

The complete sequence of the HIV-1 genome, extracted from infectious virions, has been solved to single-nucleotide resolution. [6] The HIV genome encodes a small number of viral proteins, invariably establishing cooperative associations among HIV proteins and between HIV and host proteins, to invade host cells and hijack their internal machineries. [7] HIV is different in structure from other retroviruses. The HIV virion is ~100 nm in diameter. Its innermost region consists of a cone-shaped core that includes two copies of the (positive sense) ssRNA genome, the enzymes reverse transcriptase, integrase and protease, some minor proteins, and the major core protein. [8] The genome of human immunodeficiency virus (HIV) encodes 8 viral proteins playing essential roles during the HIV life cycle. [7]
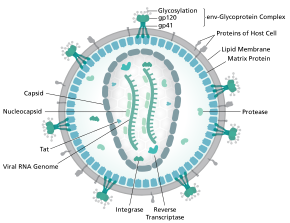
HIV-1 is composed of two copies of noncovalently linked, unspliced, positive-sense single-stranded RNA enclosed by a conical capsid composed of the viral protein p24, typical of lentiviruses. [9] [10] The two RNAs are often identical, yet they are not independent, but form a compact dimer within the virion. [11] Several reasons as for why two copies of RNA are packaged rather than just one have been proposed, including probably a combination of these advantages: One advantage is that the two copies of RNA strands are vital in contributing to HIV-1 recombination, which occurs during reverse transcription of viral replication, thus increasing genetic diversity. [11] Another advantage is that having two copies of RNA would allow the reverse transcriptase to switch templates when encountering a break in the viral RNA, thus completing the reverse transcription without loss of genetic information. [11] Yet another reason is that the dimeric nature of the RNA genome of the virus may play a structural role in viral replication. [11] The containment of two copies of single-stranded RNA within a virion but the production of only a single DNA provirus is called pseudodiploidy. [12] The RNA component is 9749 nucleotides long [13] [14] and bears a 5’ cap (Gppp), a 3’ poly(A) tail, and many open reading frames (ORFs). [15] Viral structural proteins are encoded by long ORFs, whereas smaller ORFs encode regulators of the viral life cycle: attachment, membrane fusion, replication, and assembly. [15]
The single-strand RNA is tightly bound to p7 nucleocapsid proteins, late assembly protein p6, and enzymes essential to the development of the virion, such as reverse transcriptase and integrase. Lysine tRNA is the primer of the magnesium-dependent reverse transcriptase. [9] The nucleocapsid associates with the genomic RNA (one molecule per hexamer) and protects the RNA from digestion by nucleases. Also enclosed within the virion particle are Vif, Vpr, Nef, and viral protease.[ citation needed ] The envelope of the virion is formed by a plasma membrane of host cell origin, which is supported by a matrix composed of the viral p17 protein, ensuring the integrity of the virion particle. At the surface of the virion can be found a limited number of the envelope glycoprotein (Env) of HIV, a trimer formed by heterodimers of gp120 and gp41. Env is responsible for binding to its primary host receptor, CD4, and its co-receptor (mainly CCR5 or CXCR4), leading to viral entry into its target cell. [16]
As the only proteins on the surface of the virus, the envelope glycoproteins (gp120 and gp41) are the major targets for HIV vaccine efforts. [17] Over half of the mass of the trimeric envelope spike is N-linked glycans. The density is high as the glycans shield underlying viral protein from neutralisation by antibodies. This is one of the most densely glycosylated molecules known and the density is sufficiently high to prevent the normal maturation process of glycans during biogenesis in the endoplasmic reticulum and Golgi apparatus. [18] [19] The majority of the glycans are therefore stalled as immature 'high-mannose' glycans not normally present on secreted or cell surface human glycoproteins. [20] The unusual processing and high density means that almost all broadly neutralising antibodies that have so far been identified (from a subset of patients that have been infected for many months to years) bind to or, are adapted to cope with, these envelope glycans. [21]
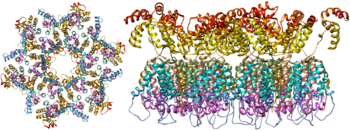
The molecular structure of the viral spike has now been determined by X-ray crystallography [22] and cryo-electron microscopy. [23] These advances in structural biology were made possible due to the development of stable recombinant forms of the viral spike by the introduction of an intersubunit disulphide bond and an isoleucine to proline mutation in gp41. [24] The so-called SOSIP trimers not only reproduce the antigenic properties of the native viral spike but also display the same degree of immature glycans as presented on the native virus. [25] Recombinant trimeric viral spikes are promising vaccine candidates as they display less non-neutralising epitopes than recombinant monomeric gp120 which act to suppress the immune response to target epitopes. [26]