
Filoviridae is a family of single-stranded negative-sense RNA viruses in the order Mononegavirales. Two members of the family that are commonly known are Ebola virus and Marburg virus. Both viruses, and some of their lesser known relatives, cause severe disease in humans and nonhuman primates in the form of viral hemorrhagic fevers.
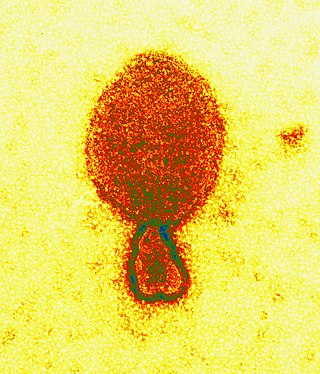
Henipavirus is a genus of negative-strand RNA viruses in the family Paramyxoviridae, order Mononegavirales containing six established species, and numerous others still under study. Henipaviruses are naturally harboured by several species of small mammals, notably pteropid fruit bats, microbats of several species, and shrews. Henipaviruses are characterised by long genomes and a wide host range. Their recent emergence as zoonotic pathogens capable of causing illness and death in domestic animals and humans is a cause of concern.

The genus Ebolavirus is a virological taxon included in the family Filoviridae, order Mononegavirales. The members of this genus are called ebolaviruses, and encode their genome in the form of single-stranded negative-sense RNA. The six known virus species are named for the region where each was originally identified: Bundibugyo ebolavirus, Reston ebolavirus, Sudan ebolavirus, Taï Forest ebolavirus, Zaire ebolavirus, and Bombali ebolavirus. The last is the most recent species to be named and was isolated from Angolan free-tailed bats in Sierra Leone. Each species of the genus Ebolavirus has one member virus, and four of these cause Ebola virus disease (EVD) in humans, a type of hemorrhagic fever having a very high case fatality rate. The Reston virus has caused EVD in other primates. Zaire ebolavirus has the highest mortality rate of the ebolaviruses and is responsible for the largest number of outbreaks of the six known species of the genus, including the 1976 Zaire outbreak and the outbreak with the most deaths (2014).
Rabies virus, scientific name Rabies lyssavirus, is a neurotropic virus that causes rabies in animals, including humans. It can cause violence, hydrophobia, and fever. Rabies transmission can also occur through the saliva of animals and less commonly through contact with human saliva. Rabies lyssavirus, like many rhabdoviruses, has an extremely wide host range. In the wild it has been found infecting many mammalian species, while in the laboratory it has been found that birds can be infected, as well as cell cultures from mammals, birds, reptiles and insects. Rabies is reported in more than 150 countries and on all continents except Antarctica. The main burden of disease is reported in Asia and Africa, but some cases have been reported also in Europe in the past 10 years, especially in returning travellers.

The M1 protein is a matrix protein of the influenza virus. It forms a coat inside the viral envelope. This is a bifunctional membrane/RNA-binding protein that mediates the encapsidation of nucleoprotein cores into the membrane envelope. It is therefore required that M1 binds both membrane and RNA simultaneously.
Group-specific antigen, or gag, is the polyprotein that contains the core structural proteins of an Ortervirus. It was named as such because scientists used to believe it was antigenic. Now it is known that it makes up the inner shell, not the envelope exposed outside. It makes up all the structural units of viral conformation and provides supportive framework for mature virion.

Membrane fusion proteins are proteins that cause fusion of biological membranes. Membrane fusion is critical for many biological processes, especially in eukaryotic development and viral entry. Fusion proteins can originate from genes encoded by infectious enveloped viruses, ancient retroviruses integrated into the host genome, or solely by the host genome. Post-transcriptional modifications made to the fusion proteins by the host, namely addition and modification of glycans and acetyl groups, can drastically affect fusogenicity.

Tumor susceptibility gene 101, also known as TSG101, is a human gene that encodes for a cellular protein of the same name.

Interferon-induced transmembrane protein 1 is a protein that in humans is encoded by the IFITM1 gene. IFITM1 has also recently been designated CD225. This protein has several additional names: fragilis, IFI17 [interferon-induced protein 17], 9-27 [Interferon-inducible protein 9-27] and Leu13.

Tetherin, also known as bone marrow stromal antigen 2, is a lipid raft associated protein that in humans is encoded by the BST2 gene. In addition, tetherin has been designated as CD317. This protein is constitutively expressed in mature B cells, plasma cells and plasmacytoid dendritic cells, and in many other cells, it is only expressed as a response to stimuli from IFN pathway.
The endosomal sorting complexes required for transport (ESCRT) machinery is made up of cytosolic protein complexes, known as ESCRT-0, ESCRT-I, ESCRT-II, and ESCRT-III. Together with a number of accessory proteins, these ESCRT complexes enable a unique mode of membrane remodeling that results in membranes bending/budding away from the cytoplasm. These ESCRT components have been isolated and studied in a number of organisms including yeast and humans. A eukaryotic signature protein, the machinery is found in all eukaryotes and some archaea.
Retroviral matrix proteins are components of envelope-associated capsids of retroviruses. These proteins line the inner surface of viral envelopes and are associated with viral membranes.
The species Lloviu cuevavirus is the taxonomic home of a virus that forms filamentous virion, Lloviu virus (LLOV). The species is included in the genus Cuevavirus. LLOV is a distant relative of the commonly known Ebola virus and Marburg virus.
The species Taï Forest ebolavirus is a virological taxon included in the genus Ebolavirus, family Filoviridae, order Mononegavirales. The species has a single virus member, Taï Forest virus (TAFV). The members of the species are called Taï Forest ebolaviruses.
The species Sudan ebolavirus is a virological taxon included in the genus Ebolavirus, family Filoviridae, order Mononegavirales. The species has a single virus member, Sudan virus (SUDV). The members of the species are called Sudan ebolaviruses. It was discovered in 1977 and causes Ebola clinically indistinguishable from the ebola Zaire strain, but is less transmissible than it. Unlike with ebola Zaire there is no vaccine available.

The family of vesiculovirus matrix proteins consists of several matrix proteins of the vesicular stomatitis virus, also known as VSIV or VSV. The matrix (M) protein of the virus causes many of the cytopathic effects of VSV, including an inhibition of host gene expression and the induction of cell rounding. It has been shown that M protein also induces apoptosis in the absence of other viral components. It is thought that the activation of apoptotic pathways causes the inhibition of host gene expression and cell rounding by M protein.

Ebola, also known as Ebola virus disease (EVD) and Ebola hemorrhagic fever (EHF), is a viral hemorrhagic fever in humans and other primates, caused by ebolaviruses. Symptoms typically start anywhere between two days and three weeks after infection. The first symptoms are usually fever, sore throat, muscle pain, and headaches. These are usually followed by vomiting, diarrhoea, rash and decreased liver and kidney function, at which point some people begin to bleed both internally and externally. It kills between 25% and 90% of those infected – about 50% on average. Death is often due to shock from fluid loss, and typically occurs between six and 16 days after the first symptoms appear. Early treatment of symptoms increases the survival rate considerably compared to late start. An Ebola vaccine was approved by the US FDA in December 2019.

Zaire ebolavirus, more commonly known as Ebola virus, is one of six known species within the genus Ebolavirus. Four of the six known ebolaviruses, including EBOV, cause a severe and often fatal hemorrhagic fever in humans and other mammals, known as Ebola virus disease (EVD). Ebola virus has caused the majority of human deaths from EVD, and was the cause of the 2013–2016 epidemic in western Africa, which resulted in at least 28,646 suspected cases and 11,323 confirmed deaths.
Ebola viral protein 24 (eVP24) is considered a multifunctional secondary matrix protein present in viral particles. The broad roles eVP24 performs involve the formation of fully functional and infectious viral particles, promotion of filamentous nucleocapsid formation, mediation of host responses to infection, and suppression of the host innate immune system. It has been noted that eVP24 function can overlap with that of two other viral proteins; eVP40 matrix protein which functions in virus budding, and eVP35 which is also associated with immune suppression.

Fadila Bouamr is a French-American virologist researching the molecular mechanisms that govern the assembly and egress of an infectious HIV-1 and the proteins involved in these processes. She is chief of the viral budding unit at the National Institute of Allergy and Infectious Diseases.














