
Rhabdoviridae is a family of negative-strand RNA viruses in the order Mononegavirales. Vertebrates, invertebrates, plants, fungi and protozoans serve as natural hosts. Diseases associated with member viruses include rabies encephalitis caused by the rabies virus, and flu-like symptoms in humans caused by vesiculoviruses. The name is derived from Ancient Greek rhabdos, meaning rod, referring to the shape of the viral particles. The family has 40 genera, most assigned to three subfamilies.

Influenza hemagglutinin (HA) or haemagglutinin[p] is a homotrimeric glycoprotein found on the surface of influenza viruses and is integral to its infectivity.

Orthomyxoviridae is a family of negative-sense RNA viruses. It includes seven genera: Alphainfluenzavirus, Betainfluenzavirus, Gammainfluenzavirus, Deltainfluenzavirus, Isavirus, Thogotovirus, and Quaranjavirus. The first four genera contain viruses that cause influenza in birds and mammals, including humans. Isaviruses infect salmon; the thogotoviruses are arboviruses, infecting vertebrates and invertebrates. The Quaranjaviruses are also arboviruses, infecting vertebrates (birds) and invertebrates (arthropods).

The mumps virus (MuV) is the virus that causes mumps. MuV contains a single-stranded, negative-sense genome made of ribonucleic acid (RNA). Its genome is about 15,000 nucleotides in length and contains seven genes that encode nine proteins. The genome is encased by a capsid that is in turn surrounded by a viral envelope. MuV particles, called virions, are pleomorphic in shape and vary in size from 100 to 600 nanometers in diameter. One serotype and twelve genotypes that vary in their geographic distribution are recognized. Humans are the only natural host of the mumps virus.

Rubella virus (RuV) is the pathogenic agent of the disease rubella, transmitted only between humans via the respiratory route, and is the main cause of congenital rubella syndrome when infection occurs during the first weeks of pregnancy.
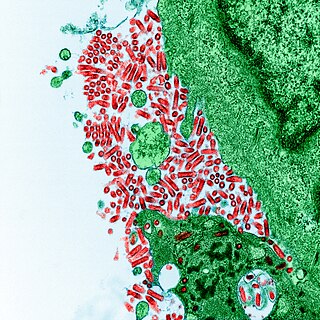
Rabies virus, scientific name Rabies lyssavirus, is a neurotropic virus that causes rabies in humans and animals. Rabies transmission can occur through the saliva of animals and less commonly through contact with human saliva. Rabies lyssavirus, like many rhabdoviruses, has an extremely wide host range. In the wild it has been found infecting many mammalian species, while in the laboratory it has been found that birds can be infected, as well as cell cultures from mammals, birds, reptiles and insects. Rabies is reported in more than 150 countries on all continents, with the exclusion of Antarctica. The main burden of disease is reported in Asia and Africa, but some cases have been reported also in Europe in the past 10 years, especially in returning travellers.
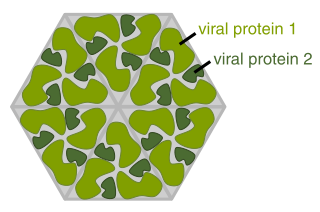
A viral protein is both a component and a product of a virus. Viral proteins are grouped according to their functions, and groups of viral proteins include structural proteins, nonstructural proteins, regulatory proteins, and accessory proteins. Viruses are non-living and do not have the means to reproduce on their own, instead depending on their host cell's resources in order to reproduce. Thus, viruses do not code for many of their own viral proteins, and instead use the host cell's machinery to produce the viral proteins they require for replication.
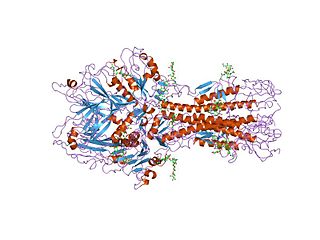
Hemagglutinin esterase (HEs) is a glycoprotein that certainenveloped viruses possess and use as an invading mechanism. HEs helps in the attachment and destruction of certain sialic acid receptors that are found on the host cell surface. Viruses that possess HEs include influenza C virus, toroviruses, and coronaviruses of the subgenus Embecovirus. HEs is a dimer transmembrane protein consisting of two monomers, each monomer is made of three domains. The three domains are: membrane fusion, esterase, and receptor binding domains.
The genome and proteins of HIV have been the subject of extensive research since the discovery of the virus in 1983. "In the search for the causative agent, it was initially believed that the virus was a form of the Human T-cell leukemia virus (HTLV), which was known at the time to affect the human immune system and cause certain leukemias. However, researchers at the Pasteur Institute in Paris isolated a previously unknown and genetically distinct retrovirus in patients with AIDS which was later named HIV." Each virion comprises a viral envelope and associated matrix enclosing a capsid, which itself encloses two copies of the single-stranded RNA genome and several enzymes. The discovery of the virus itself occurred two years following the report of the first major cases of AIDS-associated illnesses.

The Matrix-2 (M2) protein is a proton-selective viroporin, integral in the viral envelope of the influenza A virus. The channel itself is a homotetramer, where the units are helices stabilized by two disulfide bonds, and is activated by low pH. The M2 protein is encoded on the seventh RNA segment together with the M1 protein. Proton conductance by the M2 protein in influenza A is essential for viral replication.
Rous sarcoma virus (RSV) is a retrovirus and is the first oncovirus to have been described. It causes sarcoma in chickens.
Group-specific antigen, or gag, is the polyprotein that contains the core structural proteins of an Ortervirus. It was named as such because scientists used to believe it was antigenic. Now it is known that it makes up the inner shell, not the envelope exposed outside. It makes up all the structural units of viral conformation and provides supportive framework for mature virion.
Env is a viral gene that encodes the protein forming the viral envelope. The expression of the env gene enables retroviruses to target and attach to specific cell types, and to infiltrate the target cell membrane.

Viral neuraminidase is a type of neuraminidase found on the surface of influenza viruses that enables the virus to be released from the host cell. Neuraminidases are enzymes that cleave sialic acid groups from glycoproteins. Neuraminidase inhibitors are antiviral agents that inhibit influenza viral neuraminidase activity and are of major importance in the control of influenza.
Retroviral matrix proteins are components of envelope-associated capsids of retroviruses. These proteins line the inner surface of viral envelopes and are associated with viral membranes.
Mason-Pfizer monkey virus (M-PMV), formerly Simian retrovirus (SRV), is a species of retroviruses that usually infect and cause a fatal immune deficiency in Asian macaques. The ssRNA virus appears sporadically in mammary carcinoma of captive macaques at breeding facilities which expected as the natural host, but the prevalence of this virus in feral macaques remains unknown. M-PMV was transmitted naturally by virus-containing body fluids, via biting, scratching, grooming, and fighting. Cross contaminated instruments or equipment (fomite) can also spread this virus among animals.

In molecular biology, VP40 is the name of a viral matrix protein. Most commonly it is found in the Ebola virus (EBOV), a type of non-segmented, negative-strand RNA virus. Ebola virus causes a severe and often fatal haemorrhagic fever in humans, known as Ebola virus disease. The virus matrix protein VP40 is a major structural protein that plays a central role in virus assembly and budding at the plasma membrane of infected cells. VP40 proteins work by associating with cellular membranes, interacting with the cytoplasmic tails of glycoproteins and binding to the ribonucleoprotein complex.
Entebbe bat virus is an infectious disease caused by a Flavivirus that is closely related to yellow fever.

Avian metaavulavirus 2, formerly Avian paramyxovirus 2, is a species of virus belonging to the family Paramyxoviridae and genus Metaavulavirus. The virus is a negative strand RNA virus containing a monopartite genome. Avian metaavulavirus 2 is one of nine species belonging to the genus Metaavulavirus. The most common serotype of Avulavirinae is serotype 1, the cause of Newcastle disease (ND). Avian metaavulavirus 2 has been known to cause disease, specifically mild respiratory infections in domestic poultry, including turkeys and chickens, and has many economic effects on egg production and poultry industries. The virus was first isolated from a strain in Yucaipa, California in 1956. Since then, other isolates of the virus have been isolated worldwide.

The first step of transcription for some negative, single-stranded RNA viruses is cap snatching, in which the first 10 to 20 residues of a host cell RNA are removed (snatched) and used as the 5′ cap and primer to initiate the synthesis of the nascent viral mRNA. The viral RNA-dependent RNA polymerase (RdRp) can then proceed to replicate the negative-sense genome from the positive-sense template. Cap-snatching also explains why some viral mRNA have 5’ terminal extensions of 10-20 nucleotides that are not encoded for in the genome. Examples of viruses that engage in cap-snatching include influenza viruses (Orthomyxoviridae), Lassa virus (Arenaviridae), hantaan virus (Hantaviridae) and rift valley fever virus (Phenuiviridae). Most viruses snatch 15-20 nucleotides except for the families Arenaviridae and Nairoviridae and the genus Thogotovirus (Orthomyxoviridae) which use a shorter strand.













