
The hepatitis C virus (HCV) is a small, enveloped, positive-sense single-stranded RNA virus of the family Flaviviridae. The hepatitis C virus is the cause of hepatitis C and some cancers such as liver cancer and lymphomas in humans.
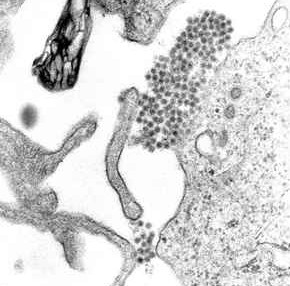
Dengue virus (DENV) is the cause of dengue fever. It is a mosquito-borne, single positive-stranded RNA virus of the family Flaviviridae; genus Flavivirus. Four serotypes of the virus have been found, and a reported fifth has yet to be confirmed, all of which can cause the full spectrum of disease. Nevertheless, scientists' understanding of dengue virus may be simplistic as, rather than distinct antigenic groups, a continuum appears to exist. This same study identified 47 strains of dengue virus. Additionally, coinfection with and lack of rapid tests for Zika virus and chikungunya complicate matters in real-world infections.
Virus-like particles (VLPs) are molecules that closely resemble viruses, but are non-infectious because they contain no viral genetic material. They can be naturally occurring or synthesized through the individual expression of viral structural proteins, which can then self assemble into the virus-like structure. Combinations of structural capsid proteins from different viruses can be used to create recombinant VLPs. Both in-vivo assembly and in-vitro assembly have been successfully shown to form virus-like particles. VLPs derived from the Hepatitis B virus (HBV) and composed of the small HBV derived surface antigen (HBsAg) were described in 1968 from patient sera. VLPs have been produced from components of a wide variety of virus families including Parvoviridae, Retroviridae, Flaviviridae, Paramyxoviridae and bacteriophages. VLPs can be produced in multiple cell culture systems including bacteria, mammalian cell lines, insect cell lines, yeast and plant cells.

Baculoviridae is a family of viruses. Arthropods, among the most studied being Lepidoptera, Hymenoptera and Diptera, serve as natural hosts. Currently, 85 species are placed in this family, assigned to four genera.
The genome and proteins of HIV (human immunodeficiency virus) have been the subject of extensive research since the discovery of the virus in 1983. "In the search for the causative agent, it was initially believed that the virus was a form of the Human T-cell leukemia virus (HTLV), which was known at the time to affect the human immune system and cause certain leukemias. However, researchers at the Pasteur Institute in Paris isolated a previously unknown and genetically distinct retrovirus in patients with AIDS which was later named HIV." Each virion comprises a viral envelope and associated matrix enclosing a capsid, which itself encloses two copies of the single-stranded RNA genome and several enzymes. The discovery of the virus itself occurred two years following the report of the first major cases of AIDS-associated illnesses.

Envelope glycoprotein GP120 is a glycoprotein exposed on the surface of the HIV envelope. It was discovered by Professors Tun-Hou Lee and Myron "Max" Essex of the Harvard School of Public Health in 1988. The 120 in its name comes from its molecular weight of 120 kDa. Gp120 is essential for virus entry into cells as it plays a vital role in attachment to specific cell surface receptors. These receptors are DC-SIGN, Heparan Sulfate Proteoglycan and a specific interaction with the CD4 receptor, particularly on helper T-cells. Binding to CD4 induces the start of a cascade of conformational changes in gp120 and gp41 that lead to the fusion of the viral membrane with the host cell membrane. Binding to CD4 is mainly electrostatic although there are van der Waals interactions and hydrogen bonds.
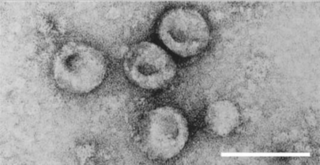
Pestivirus is a genus of viruses, in the family Flaviviridae. Viruses in the genus Pestivirus infect mammals, including members of the family Bovidae and the family Suidae. There are 11 species in this genus. Diseases associated with this genus include: hemorrhagic syndromes, abortion, and fatal mucosal disease.
The murine leukemia viruses are retroviruses named for their ability to cause cancer in murine (mouse) hosts. Some MLVs may infect other vertebrates. MLVs include both exogenous and endogenous viruses. Replicating MLVs have a positive sense, single-stranded RNA (ssRNA) genome that replicates through a DNA intermediate via the process of reverse transcription.

A viral envelope is the outermost layer of many types of viruses. It protects the genetic material in their life cycle when traveling between host cells. Not all viruses have envelopes. A viral envelope protein or E protein is a protein in the envelope, which may be acquired by the capsid from an infected host cell.

Herpes simplex virus1 and 2, also known by their taxonomical names Human alphaherpesvirus 1 and Human alphaherpesvirus 2, are two members of the human Herpesviridae family, a set of viruses that produce viral infections in the majority of humans. Both HSV-1 and HSV-2 are very common and contagious. They can be spread when an infected person begins shedding the virus.

Viral entry is the earliest stage of infection in the viral life cycle, as the virus comes into contact with the host cell and introduces viral material into the cell. The major steps involved in viral entry are shown below. Despite the variation among viruses, there are several shared generalities concerning viral entry.
Env is a viral gene that encodes the protein forming the viral envelope. The expression of the env gene enables retroviruses to target and attach to specific cell types, and to infiltrate the target cell membrane.

CD81 molecule, also known as CD81, is a protein which in humans is encoded by the CD81 gene. It is also known as 26 kDa cell surface protein, TAPA-1, and Tetraspanin-28 (Tspan-28).
NS2-3 protease is an enzyme responsible for proteolytic cleavage between NS2 and NS3, which are non-structural proteins that form part of the HCV virus particle. NS3 protease of hepatitis C virus, on the other hand, is responsible for the cleavage of non-structural protein downstream. Both of these proteases are directly involved in HCV genome replication, that is, during the viral life-cycle that leads to virus multiplication in the host that has been infected by the virus.

E2 is a viral structural protein found in the hepatitis C virus. It is present on the viral envelope and functions as a host receptor binding protein, mediating entry into host cells. It is a key target for the design of entry inhibitors and vaccine immunogens.

Nonstructural protein 2 (NS2) is a viral protein found in the hepatitis C virus. It is also produced by influenza viruses, and is alternatively known as the nuclear export protein (NEP).
A hepatitis C vaccine, a vaccine capable of protecting against the hepatitis C virus (HCV), is not yet available. Although vaccines exist for hepatitis A and hepatitis B, development of an HCV vaccine has presented challenges. No vaccine is currently available, but several vaccines are currently under development.

Hepatitis B virus (HBV) is a partially double-stranded DNA virus, a species of the genus Orthohepadnavirus and a member of the Hepadnaviridae family of viruses. This virus causes the disease hepatitis B.
Mason-Pfizer monkey virus (M-PMV), formerly Simian retrovirus (SRV), is a species of retroviruses that usually infect and cause a fatal immune deficiency in Asian macaques. The ssRNA virus appears sporadically in mammary carcinoma of captive macaques at breeding facilities which expected as the natural host, but the prevalence of this virus in feral macaques remains unknown. M-PMV was transmitted naturally by virus-containing body fluids, via biting, scratching, grooming, and fighting. Cross contaminated instruments or equipment (fomite) can also spread this virus among animals.

Nonstructural protein 5A (NS5A) inhibitors are direct acting antiviral agents (DAAs) that target viral proteins, and their development was a culmination of increased understanding of the viral life cycle combined with advances in drug discovery technology. However, their mechanism of action is complex and not fully understood. NS5A inhibitors were the focus of much attention when they emerged as a part of the first curative treatment for hepatitis C virus (HCV) infections in 2014. Favorable characteristics have been introduced through varied structural changes, and structural similarities between NS5A inhibitors that are clinically approved are readily apparent. Despite the recent introduction of numerous new antiviral drugs, resistance is still a concern and these inhibitors are therefore always used in combination with other drugs.











