Coronaviruses are a group of related RNA viruses that cause diseases in mammals and birds. In humans and birds, they cause respiratory tract infections that can range from mild to lethal. Mild illnesses in humans include some cases of the common cold, while more lethal varieties can cause SARS, MERS and COVID-19. In cows and pigs they cause diarrhea, while in mice they cause hepatitis and encephalomyelitis.

Betacoronavirus pandemicum is a species of virus consisting of many known strains. Two strains of the virus have caused outbreaks of severe respiratory diseases in humans: severe acute respiratory syndrome coronavirus 1, the cause of the 2002–2004 outbreak of severe acute respiratory syndrome (SARS), and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the cause of the pandemic of COVID-19. There are hundreds of other strains of SARSr-CoV, which are only known to infect non-human mammal species: bats are a major reservoir of many strains of SARSr-CoV; several strains have been identified in Himalayan palm civets, which were likely ancestors of SARS-CoV-1.

Coronaviridae is a family of enveloped, positive-strand RNA viruses which infect amphibians, birds, and mammals. The group includes the subfamilies Letovirinae and Orthocoronavirinae; the members of the latter are known as coronaviruses.

Human coronavirus NL63 (HCoV-NL63) is a species of coronavirus, specifically a Setracovirus from among the Alphacoronavirus genus. It was identified in late 2004 in patients in the Netherlands by Lia van der Hoek and Krzysztof Pyrc using a novel virus discovery method VIDISCA. Later on the discovery was confirmed by the researchers from Rotterdam. The virus is an enveloped, positive-sense, single-stranded RNA virus which enters its host cell by binding to ACE2. Infection with the virus has been confirmed worldwide, and has an association with many common symptoms and diseases. Associated diseases include mild to moderate upper respiratory tract infections, severe lower respiratory tract infection, croup and bronchiolitis.

Vpu is an accessory protein that in HIV is encoded by the vpu gene. Vpu stands for "Viral Protein U". The Vpu protein acts in the degradation of CD4 in the endoplasmic reticulum and in the enhancement of virion release from the plasma membrane of infected cells. Vpu induces the degradation of the CD4 viral receptor and therefore participates in the general downregulation of CD4 expression during the course of HIV infection. Vpu-mediated CD4 degradation is thought to prevent CD4-Env binding in the endoplasmic reticulum to facilitate proper Env assembly into virions. It is found in the membranes of infected cells, but not the virus particles themselves.

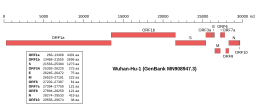
ORF7a is a gene found in coronaviruses of the Betacoronavirus genus. It expresses the Betacoronavirus NS7A protein, a type I transmembrane protein with an immunoglobulin-like protein domain. It was first discovered in SARS-CoV, the virus that causes severe acute respiratory syndrome (SARS). The homolog in SARS-CoV-2, the virus that causes COVID-19, has about 85% sequence identity to the SARS-CoV protein.

Embecovirus is a subgenus of coronaviruses in the genus Betacoronavirus. The viruses in this subgenus, unlike other coronaviruses, have a hemagglutinin esterase (HE) gene. The viruses in the subgenus were previously known as group 2a coronaviruses.

Merbecovirus is a subgenus of viruses in the genus Betacoronavirus, including the human pathogen Middle East respiratory syndrome–related coronavirus (MERS-CoV). The viruses in this subgenus were previously known as group 2c coronaviruses.
ORF3b is a gene found in coronaviruses of the subgenus Sarbecovirus, encoding a short non-structural protein. It is present in both SARS-CoV and SARS-CoV-2, though the protein product has very different lengths in the two viruses. The encoded protein is significantly shorter in SARS-CoV-2, at only 22 amino acid residues compared to 153–155 in SARS-CoV. Both the longer SARS-CoV and shorter SARS-CoV-2 proteins have been reported as interferon antagonists. It is unclear whether the SARS-CoV-2 gene expresses a functional protein.
ORF3d is a gene found in SARS-CoV-2 and at least one closely related coronavirus found in pangolins, though it is not found in other closely related viruses within the Sarbecovirus subgenus. It is 57 codons long and encodes a novel 57 amino acid residue protein of unknown function. At least two isoforms have been described, of which the shorter 33-residue form, ORF3d-2, may be more highly expressed, or even the only form expressed. It is reported to be antigenic and antibodies to the ORF3d protein occur in patients recovered from COVID-19. There is no homolog in the genome of the otherwise closely related SARS-CoV.

The envelope (E) protein is the smallest and least well-characterized of the four major structural proteins found in coronavirus virions. It is an integral membrane protein less than 110 amino acid residues long; in SARS-CoV-2, the causative agent of Covid-19, the E protein is 75 residues long. Although it is not necessarily essential for viral replication, absence of the E protein may produce abnormally assembled viral capsids or reduced replication. E is a multifunctional protein and, in addition to its role as a structural protein in the viral capsid, it is thought to be involved in viral assembly, likely functions as a viroporin, and is involved in viral pathogenesis.

The membrane (M) protein is an integral membrane protein that is the most abundant of the four major structural proteins found in coronaviruses. The M protein organizes the assembly of coronavirus virions through protein-protein interactions with other M protein molecules as well as with the other three structural proteins, the envelope (E), spike (S), and nucleocapsid (N) proteins.

The nucleocapsid (N) protein is a protein that packages the positive-sense RNA genome of coronaviruses to form ribonucleoprotein structures enclosed within the viral capsid. The N protein is the most highly expressed of the four major coronavirus structural proteins. In addition to its interactions with RNA, N forms protein-protein interactions with the coronavirus membrane protein (M) during the process of viral assembly. N also has additional functions in manipulating the cell cycle of the host cell. The N protein is highly immunogenic and antibodies to N are found in patients recovered from SARS and COVID-19.
ORF3c is a gene found in coronaviruses of the subgenus Sarbecovirus, including SARS-CoV and SARS-CoV-2. It was first identified in the SARS-CoV-2 genome and encodes a 41 amino acid non-structural protein of unknown function. It is also present in the SARS-CoV genome, but was not recognized until the identification of the SARS-CoV-2 homolog.
ORF7b is a gene found in coronaviruses of the genus Betacoronavirus, which expresses the accessory protein Betacoronavirus NS7b protein. It is a short, highly hydrophobic transmembrane protein of unknown function.

ORF8 is a gene that encodes a viral accessory protein, Betacoronavirus NS8 protein, in coronaviruses of the subgenus Sarbecovirus. It is one of the least well conserved and most variable parts of the genome. In some viruses, a deletion splits the region into two smaller open reading frames, called ORF8a and ORF8b - a feature present in many SARS-CoV viral isolates from later in the SARS epidemic, as well as in some bat coronaviruses. For this reason the full-length gene and its protein are sometimes called ORF8ab. The full-length gene, exemplified in SARS-CoV-2, encodes a protein with an immunoglobulin domain of unknown function, possibly involving interactions with the host immune system. It is similar in structure to the ORF7a protein, suggesting it may have originated through gene duplication.
ORF6 is a gene that encodes a viral accessory protein in coronaviruses of the subgenus Sarbecovirus, including SARS-CoV and SARS-CoV-2. It is not present in MERS-CoV. It is thought to reduce the immune system response to viral infection through interferon antagonism.

ORF9b is a gene that encodes a viral accessory protein in coronaviruses of the subgenus Sarbecovirus, including SARS-CoV and SARS-CoV-2. It is an overlapping gene whose open reading frame is entirely contained within the N gene, which encodes coronavirus nucleocapsid protein. The encoded protein is 97 amino acid residues long in SARS-CoV and 98 in SARS-CoV-2, in both cases forming a protein dimer.
ORF9c is an open reading frame (ORF) in coronavirus genomes of the subgenus Sarbecovirus. It is 73 codons long in the SARS-CoV-2 genome. Although it is often included in lists of Sarbecovirus viral accessory protein genes, experimental and bioinformatics evidence suggests ORF9c may not be a functional protein-coding gene.
ORF10 is an open reading frame (ORF) found in the genome of the SARS-CoV-2 coronavirus. It is 38 codons long. It is not conserved in all Sarbecoviruses. In studies prompted by the COVID-19 pandemic, ORF10 attracted research interest as one of two viral accessory protein genes not conserved between SARS-CoV and SARS-CoV-2 and was initially described as a protein-coding gene likely under positive selection. However, although it is sometimes included in lists of SARS-CoV-2 accessory genes, experimental and bioinformatics evidence suggests ORF10 is likely not a functional protein-coding gene.