
Coronaviruses are a group of related RNA viruses that cause diseases in mammals and birds. In humans and birds, they cause respiratory tract infections that can range from mild to lethal. Mild illnesses in humans include some cases of the common cold, while more lethal varieties can cause SARS, MERS and COVID-19, which is causing the ongoing pandemic. In cows and pigs they cause diarrhea, while in mice they cause hepatitis and encephalomyelitis.

Severe acute respiratory syndrome–related coronavirus is a species of virus consisting of many known strains phylogenetically related to severe acute respiratory syndrome coronavirus 1 (SARS-CoV-1) that have been shown to possess the capability to infect humans, bats, and certain other mammals. These enveloped, positive-sense single-stranded RNA viruses enter host cells by binding to the angiotensin-converting enzyme 2 (ACE2) receptor. The SARSr-CoV species is a member of the genus Betacoronavirus and of the subgenus Sarbecovirus.

Murine coronavirus (M-CoV) is a virus in the genus Betacoronavirus that infects mice. Belonging to the subgenus Embecovirus, murine coronavirus strains are enterotropic or polytropic. Enterotropic strains include mouse hepatitis virus (MHV) strains D, Y, RI, and DVIM, whereas polytropic strains, such as JHM and A59, primarily cause hepatitis, enteritis, and encephalitis. Murine coronavirus is an important pathogen in the laboratory mouse and the laboratory rat. It is the most studied coronavirus in animals other than humans, and has been used as an animal disease model for many virological and clinical studies.
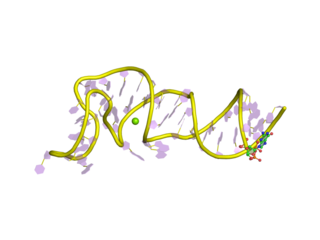
The Coronavirus 3′ stem-loop II-like motif is a secondary structure motif identified in the 3′ untranslated region (3′UTR) of astrovirus, coronavirus and equine rhinovirus genomes. Its function is unknown, but various viral 3′ UTR regions have been found to play roles in viral replication and packaging.
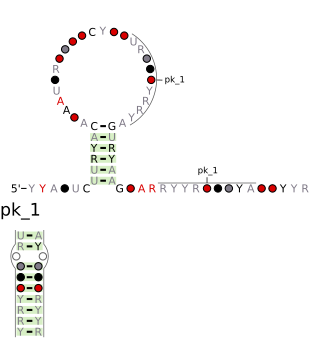
The Coronavirus 3′ UTR pseudoknot is an RNA structure found in the coronavirus genome. Coronaviruses contain 30 kb single-stranded positive-sense RNA genomes. The 3′ UTR region of these coronavirus genomes contains a conserved ~55 nucleotide pseudoknot structure which is necessary for viral genome replication. The mechanism of cis-regulation is unclear, but this element is postulated to function in the plus-strand.

In molecular biology, the coronavirus frameshifting stimulation element is a conserved stem-loop of RNA found in coronaviruses that can promote ribosomal frameshifting. Such RNA molecules interact with a downstream region to form a pseudoknot structure; the region varies according to the virus but pseudoknot formation is known to stimulate frameshifting. In the classical situation, a sequence 32 nucleotides downstream of the stem is complementary to part of the loop. In other coronaviruses, however, another stem-loop structure around 150 nucleotides downstream can interact with members of this family to form kissing stem-loops and stimulate frameshifting.

The Infectious bronchitis virus D-RNA is an RNA element known as defective RNA or D-RNA. This element is thought to be essential for viral replication and efficient packaging of avian infectious bronchitis virus (IBV) particles.
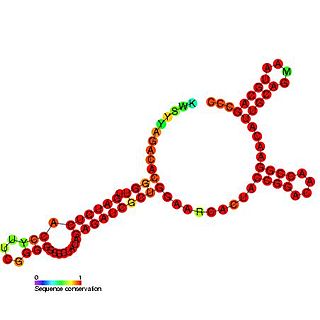
Tombusvirus 3′ UTR is an important cis-regulatory region of the Tombus virus genome.
NSP1 (NS53), the product of rotavirus gene 5, is a nonstructural RNA-binding protein that contains a cysteine-rich region and is a component of early replication intermediates. RNA-folding predictions suggest that this region of the NSP1 mRNA can interact with itself, producing a stem-loop structure similar to that found near the 5'-terminus of the NSP1 mRNA.
Putative transmembrane domain more commonly known as Non-structural Protein 6 (NSP6) is one of the two non-structural proteins that gene 11 in rotavirus encodes for alongside NSP5. NSP6 is composed of six transmembrane domains and a C terminal tail. In contrast to the other rotavirus non-structural proteins, NSP6 was found to have a high rate of turnover, being completely degraded within 2 hours of synthesis. NSP6 was found to be a sequence-independent nucleic acid binding protein, with similar affinities for ssRNA and dsRNA

Betacoronavirus is one of four genera of coronaviruses. Member viruses are enveloped, positive-strand RNA viruses that infect mammals. The natural reservoir for betacoronaviruses are bats and rodents. Rodents are the reservoir for the subgenus Embecovirus, while bats are the reservoir for the other subgenera.

Human coronavirus OC43 (HCoV-OC43) is a member of the species Betacoronavirus 1, which infects humans and cattle. The infecting coronavirus is an enveloped, positive-sense, single-stranded RNA virus that enters its host cell by binding to the N-acetyl-9-O-acetylneuraminic acid receptor. OC43 is one of seven coronaviruses known to infect humans. It is one of the viruses responsible for the common cold and may have been responsible for the 1889–1890 pandemic. It has, like other coronaviruses from genus Betacoronavirus, subgenus Embecovirus, an additional shorter spike protein called hemagglutinin-esterase (HE).

Alphacoronaviruses (Alpha-CoV) are members of the first of the four genera of coronaviruses. They are positive-sense, single-stranded RNA viruses that infect mammals, including humans. They have spherical virions with club-shaped surface projections formed by trimers of the spike protein, and a viral envelope.

Positive-strand RNA viruses are a group of related viruses that have positive-sense, single-stranded genomes made of ribonucleic acid. The positive-sense genome can act as messenger RNA (mRNA) and can be directly translated into viral proteins by the host cell's ribosomes. Positive-strand RNA viruses encode an RNA-dependent RNA polymerase (RdRp) which is used during replication of the genome to synthesize a negative-sense antigenome that is then used as a template to create a new positive-sense viral genome.

Embecovirus is a subgenus of coronaviruses in the genus Betacoronavirus. The viruses in this subgenus, unlike other coronaviruses, have a hemagglutinin esterase (HE) gene. The viruses in the subgenus were previously known as group 2a coronaviruses.

Merbecovirus is a subgenus of viruses in the genus Betacoronavirus, including the human pathogen Middle East respiratory syndrome–related coronavirus (MERS-CoV). The viruses in this subgenus were previously known as group 2c coronaviruses.
Coronavirus genomes are positive-sense single-stranded RNA molecules with an untranslated region (UTR) at the 5′ end which is called the 5′ UTR. The 5′ UTR is responsible for important biological functions, such as viral replication, transcription and packaging. The 5′ UTR has a conserved RNA secondary structure but different Coronavirus genera have different structural features described below.
Coronavirus genomes are positive-sense single-stranded RNA molecules with an untranslated region (UTR) at the 3′ end which is called the 3′ UTR. The 3′ UTR is responsible for important biological functions, such as viral replication. The 3′ UTR has a conserved RNA secondary structure but different Coronavirus genera have different structural features described below.

The nucleocapsid (N) protein is a protein that packages the positive-sense RNA genome of coronaviruses to form ribonucleoprotein structures enclosed within the viral capsid. The N protein is the most highly expressed of the four major coronavirus structural proteins. In addition to its interactions with RNA, N forms protein-protein interactions with the coronavirus membrane protein (M) during the process of viral assembly. N also has additional functions in manipulating the cell cycle of the host cell. The N protein is highly immunogenic and antibodies to N are found in patients recovered from SARS and Covid-19.
ORF1ab refers collectively to two open reading frames (ORFs), ORF1a and ORF1b, that are conserved in the genomes of nidoviruses, a group of viruses that includes coronaviruses. The genes express large polyproteins that undergo proteolysis to form several nonstructural proteins with various functions in the viral life cycle, including proteases and the components of the replicase-transcriptase complex (RTC). Together the two ORFs are sometimes referred to as the replicase gene. They are related by a programmed ribosomal frameshift that allows the ribosome to continue translating past the stop codon at the end of ORF1a, in a -1 reading frame. The resulting polyproteins are known as pp1a and pp1ab.













