Related Research Articles

Interferons are a group of signaling proteins made and released by host cells in response to the presence of several viruses. In a typical scenario, a virus-infected cell will release interferons causing nearby cells to heighten their anti-viral defenses.
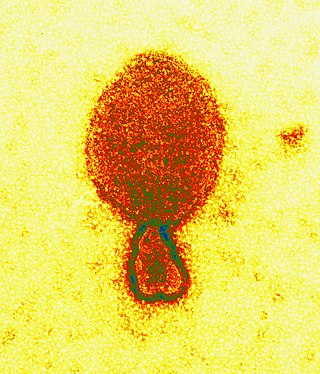
Henipavirus is a genus of negative-strand RNA viruses in the family Paramyxoviridae, order Mononegavirales containing six established species, and numerous others still under study. Henipaviruses are naturally harboured by several species of small mammals, notably pteropid fruit bats, microbats of several species, and shrews. Henipaviruses are characterised by long genomes and a wide host range. Their recent emergence as zoonotic pathogens capable of causing illness and death in domestic animals and humans is a cause of concern.
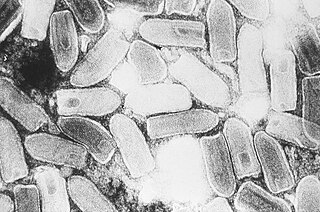
Indiana vesiculovirus, formerly Vesicular stomatitis Indiana virus is a virus in the family Rhabdoviridae; the well-known Rabies lyssavirus belongs to the same family. VSIV can infect insects, cattle, horses and pigs. It has particular importance to farmers in certain regions of the world where it infects cattle. This is because its clinical presentation is identical to the very important foot and mouth disease virus.
The JAK-STAT signaling pathway is a chain of interactions between proteins in a cell, and is involved in processes such as immunity, cell division, cell death, and tumour formation. The pathway communicates information from chemical signals outside of a cell to the cell nucleus, resulting in the activation of genes through the process of transcription. There are three key parts of JAK-STAT signalling: Janus kinases (JAKs), signal transducer and activator of transcription proteins (STATs), and receptors. Disrupted JAK-STAT signalling may lead to a variety of diseases, such as skin conditions, cancers, and disorders affecting the immune system.

Interferon gamma (IFN-γ) is a dimerized soluble cytokine that is the only member of the type II class of interferons. The existence of this interferon, which early in its history was known as immune interferon, was described by E. F. Wheelock as a product of human leukocytes stimulated with phytohemagglutinin, and by others as a product of antigen-stimulated lymphocytes. It was also shown to be produced in human lymphocytes. or tuberculin-sensitized mouse peritoneal lymphocytes challenged with Mantoux test (PPD); the resulting supernatants were shown to inhibit growth of vesicular stomatitis virus. Those reports also contained the basic observation underlying the now widely employed IFN-γ release assay used to test for tuberculosis. In humans, the IFN-γ protein is encoded by the IFNG gene.

A viral envelope is the outermost layer of many types of viruses. It protects the genetic material in their life cycle when traveling between host cells. Not all viruses have envelopes. A viral envelope protein or E protein is a protein in the envelope, which may be acquired by the capsid from an infected host cell.
The interferon-α/β receptor (IFNAR) is a virtually ubiquitous membrane receptor which binds endogenous type I interferon (IFN) cytokines. Endogenous human type I IFNs include many subtypes, such as interferons-α, -β, -ε, -κ, -ω, and -ζ.

Signal transducer and activator of transcription 1 (STAT1) is a transcription factor which in humans is encoded by the STAT1 gene. It is a member of the STAT protein family.

Antibody-dependent enhancement (ADE), sometimes less precisely called immune enhancement or disease enhancement, is a phenomenon in which binding of a virus to suboptimal antibodies enhances its entry into host cells, followed by its replication. The suboptimal antibodies can result from natural infection or from vaccination. ADE may cause enhanced respiratory disease, but is not limited to respiratory disease. It has been observed in HIV, RSV virus and Dengue virus and is monitored for in vaccine development.

Signal transducer and activator of transcription 2 is a protein that in humans is encoded by the STAT2 gene. It is a member of the STAT protein family. This protein is critical to the biological response of type I interferons (IFNs). STAT2 sequence identity between mouse and human is only 68%.

Importin subunit alpha-1 is a protein that in humans is encoded by the KPNA2 gene.

Importin subunit alpha-7 is a protein that in humans is encoded by the KPNA6 gene.

Importin subunit alpha-3, also known as karyopherin subunit alpha-4, is a protein that in humans is encoded by the KPNA4 gene.

Importin subunit alpha-6 is a protein that in humans is encoded by the KPNA5 gene.

Tetherin, also known as bone marrow stromal antigen 2, is a lipid raft associated protein that in humans is encoded by the BST2 gene. In addition, tetherin has been designated as CD317. This protein is constitutively expressed in mature B cells, plasma cells and plasmacytoid dendritic cells, and in many other cells, it is only expressed as a response to stimuli from IFN pathway.
Importin alpha, or karyopherin alpha refers to a class of adaptor proteins that are involved in the import of proteins into the cell nucleus. They are a sub-family of karyopherin proteins.

Zaire ebolavirus, more commonly known as Ebola virus, is one of six known species within the genus Ebolavirus. Four of the six known ebolaviruses, including EBOV, cause a severe and often fatal hemorrhagic fever in humans and other mammals, known as Ebola virus disease (EVD). Ebola virus has caused the majority of human deaths from EVD, and was the cause of the 2013–2016 epidemic in western Africa, which resulted in at least 28,646 suspected cases and 11,323 confirmed deaths.
The cGAS–STING pathway is a component of the innate immune system that functions to detect the presence of cytosolic DNA and, in response, trigger expression of inflammatory genes that can lead to senescence or to the activation of defense mechanisms. DNA is normally found in the nucleus of the cell. Localization of DNA to the cytosol is associated with tumorigenesis, viral infection, and invasion by some intracellular bacteria. The cGAS – STING pathway acts to detect cytosolic DNA and induce an immune response.
ORF6 is a gene that encodes a viral accessory protein in coronaviruses of the subgenus Sarbecovirus, including SARS-CoV and SARS-CoV-2. It is not present in MERS-CoV. It is thought to reduce the immune system response to viral infection through interferon antagonism.
Endothelial cell tropism or endotheliotropism is a type of tissue tropism or host tropism that characterizes an pathogen's ability to recognize and infect an endothelial cell. Pathogens, such as viruses, can target a specific tissue type or multiple tissue types. Like other cells, the endothelial cell possesses several features that supports a productive viral infection a cell including, cell surface receptors, immune responses, and other virulence factors. Endothelial cells are found in various tissue types such as in the capillaries, veins, and arteries in the human body. As endothelial cells line these blood vessels and critical networks that extend access to various human organ systems, the virus entry into these cells can be detrimental to virus spread across the host system and affect clinical course of disease. Understanding the mechanisms of how viruses attach, enter, and control endothelial functions and host responses inform infectious disease understanding and medical countermeasures.
References
- 1 2 3 Shabman RS, Gulcicek EE, Stone KL, Basler CF (November 2011). "The Ebola virus VP24 protein prevents hnRNP C1/C2 binding to karyopherin α1 and partially alters its nuclear import". The Journal of Infectious Diseases. 204 Suppl 3: S904-10. doi:10.1093/infdis/jir323. PMC 3189985 . PMID 21987768.
- 1 2 Han Z, Boshra H, Sunyer JO, Zwiers SH, Paragas J, Harty RN (February 2003). "Biochemical and functional characterization of the Ebola virus VP24 protein: implications for a role in virus assembly and budding". Journal of Virology. 77 (3): 1793–800. doi:10.1128/JVI.77.3.1793-1800.2003. PMC 140957 . PMID 12525613.
- 1 2 3 4 5 6 Xu W, Edwards MR, Borek DM, Feagins AR, Mittal A, Alinger JB, et al. (August 2014). "Ebola virus VP24 targets a unique NLS binding site on karyopherin alpha 5 to selectively compete with nuclear import of phosphorylated STAT1". Cell Host & Microbe. 16 (2): 187–200. doi:10.1016/j.chom.2014.07.008. PMC 4188415 . PMID 25121748.
- ↑ Pourrut X, Kumulungui B, Wittmann T, Moussavou G, Délicat A, Yaba P, Nkoghe D, Gonzalez JP, Leroy EM (June 2005). "The natural history of Ebola virus in Africa". Microbes and Infection. 7 (7–8): 1005–14. doi: 10.1016/j.micinf.2005.04.006 . PMID 16002313.
- ↑ Sullivan N, Yang ZY, Nabel GJ (September 2003). "Ebola virus pathogenesis: implications for vaccines and therapies". Journal of Virology. 77 (18): 9733–7. doi:10.1128/JVI.77.18.9733-9737.2003. PMC 224575 . PMID 12941881.
- 1 2 3 4 5 6 Daugherty MD, Malik HS (August 2014). "How a virus blocks a cellular emergency access lane to the nucleus, STAT!". Cell Host & Microbe. 16 (2): 150–152. doi: 10.1016/j.chom.2014.07.013 . PMID 25121743.
- ↑ Yoshimoto T, Okada K, Morishima N, Kamiya S, Owaki T, Asakawa M, Iwakura Y, Fukai F, Mizuguchi J (August 2004). "Induction of IgG2a class switching in B cells by IL-27". Journal of Immunology. 173 (4): 2479–85. doi: 10.4049/jimmunol.173.4.2479 . PMID 15294962.
- ↑ Stephanou A, Brar BK, Knight RA, Latchman DS (March 2000). "Opposing actions of STAT-1 and STAT-3 on the Bcl-2 and Bcl-x promoters". Cell Death and Differentiation. 7 (3): 329–30. doi: 10.1038/sj.cdd.4400656 . PMID 10866494.
- ↑ Lee CK, Smith E, Gimeno R, Gertner R, Levy DE (February 2000). "STAT1 affects lymphocyte survival and proliferation partially independent of its role downstream of IFN-gamma". Journal of Immunology. 164 (3): 1286–92. doi: 10.4049/jimmunol.164.3.1286 . PMID 10640742.
- ↑ Wilson JA, Bray M, Bakken R, Hart MK (August 2001). "Vaccine potential of Ebola virus VP24, VP30, VP35, and VP40 proteins". Virology. 286 (2): 384–90. doi: 10.1006/viro.2001.1012 . PMID 11485406.