| Tat | |||||||||
|---|---|---|---|---|---|---|---|---|---|
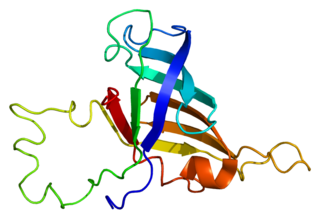
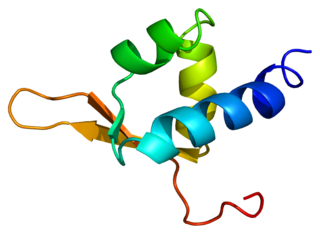
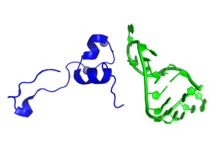 Representation of a fragment of the Tat protein (blue) from HIV along with two Zn molecules shown as grey spheres, in complex with HIV TAR RNA (green). Prepared using the data published under PDB code: 6CYT. | |||||||||
| Identifiers | |||||||||
| Symbol | Tat | ||||||||
| Pfam | PF00539 | ||||||||
| InterPro | IPR001831 | ||||||||
| PROSITE | PDOC00836 | ||||||||
| SCOP2 | 1tvs / SCOPe / SUPFAM | ||||||||
| |||||||||
In molecular biology, Tat is a protein that is encoded for by the tat gene in HIV-1. [1] [2] Tat is a regulatory protein that drastically enhances the efficiency of viral transcription. [2] Tat stands for "Trans-Activator of Transcription". The protein consists of between 86 and 101 amino acids depending on the subtype. [3] Tat vastly increases the level of transcription of the HIV dsDNA. Before Tat is present, a small number of RNA transcripts will be made, which allow the Tat protein to be produced. Tat then binds to cellular factors and mediates their phosphorylation, resulting in increased transcription of all HIV genes, [4] providing a positive feedback cycle. This in turn allows HIV to have an explosive response once a threshold amount of Tat is produced, a useful tool for defeating the body's response.
Contents
Tat also appears to play a more direct role in the HIV disease process. The protein is released by infected cells in culture, and is found in the blood of HIV-1 infected patients. [5]
It can be absorbed by cells that are not infected with HIV, and can act directly as a toxin producing cell death via apoptosis in uninfected "bystander" T cells, assisting in progression toward AIDS. [6]
By antagonizing the CXCR4 receptor, Tat also appears to selectively encourage the reproduction of less virulent M-tropic (macrophage-tropic) strains of HIV (which use the CCR5 receptor) early in the course of infection, allowing the more rapidly pathogenic T-tropic (T-cell-tropic) strains (which use the CXCR4 receptor) to emerge later after mutating from M-tropic strains. [5]