Related Research Articles
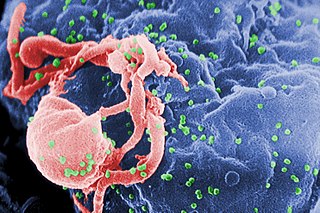
The human immunodeficiency viruses (HIV) are two species of Lentivirus that infect humans. Over time they cause acquired immunodeficiency syndrome (AIDS), a condition in which progressive failure of the immune system allows life-threatening opportunistic infections and cancers to thrive. Without treatment, average survival time after infection with HIV is estimated to be 9 to 11 years, depending on the HIV subtype. In most cases, HIV is a sexually transmitted infection and occurs by contact with or transfer of blood, pre-ejaculate, semen, and vaginal fluids. Research has shown that HIV is untransmittable through condomless sexual intercourse if the HIV-positive partner has a consistently undetectable viral load. Non-sexual transmission can occur from an infected mother to her infant during pregnancy, during childbirth by exposure to her blood or vaginal fluid, and through breast milk. Within these bodily fluids, HIV is present as both free virus particles and virus within infected immune cells.
Simian immunodeficiency virus (SIV) is a species of retrovirus that cause persistent infections in at least 45 species of African non-human primates. Based on analysis of strains found in four species of monkeys from Bioko Island, which was isolated from the mainland by rising sea levels about 11,000 years ago, it has been concluded that SIV has been present in monkeys and apes for at least 32,000 years, and probably much longer.
Lentivirus is a genus of retroviruses that cause chronic and deadly diseases characterized by long incubation periods, in the human and other mammalian species. The best known lentivirus is the human immunodeficiency virus (HIV), which causes AIDS. Lentiviruses are also hosted in apes, cows, goats, horses, cats, and sheep. Recently, lentiviruses have been found in monkeys, lemurs, Malayan flying lemur, rabbits, and ferrets. Lentiviruses and their hosts have worldwide distribution. Lentiviruses can integrate a significant amount of viral cDNA into the DNA of the host cell and can efficiently infect nondividing cells, so they are one of the most efficient methods of gene delivery. Lentiviruses can become endogenous (ERV), integrating their genome into the host germline genome, so that the virus is henceforth inherited by the host's descendants.
The genome and proteins of HIV have been the subject of extensive research since the discovery of the virus in 1983. "In the search for the causative agent, it was initially believed that the virus was a form of the Human T-cell leukemia virus (HTLV), which was known at the time to affect the human immune system and cause certain leukemias. However, researchers at the Pasteur Institute in Paris isolated a previously unknown and genetically distinct retrovirus in patients with AIDS which was later named HIV." Each virion comprises a viral envelope and associated matrix enclosing a capsid, which itself encloses two copies of the single-stranded RNA genome and several enzymes. The discovery of the virus itself occurred two years following the report of the first major cases of AIDS-associated illnesses.

Viral infectivity factor, or Vif, is an accessory protein found in HIV and other lentiviruses. Its role is to disrupt the antiviral activity of the human enzyme APOBEC by targeting it for ubiquitination and cellular degradation. APOBEC is a cytidine deaminase enzyme that mutates viral nucleic acids.
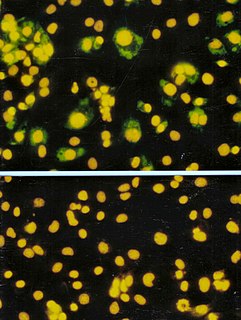
A viroplasm, sometimes called 'virus factory' or 'virus inclusion'. is an inclusion body in a cell where viral replication and assembly occurs. They may be thought of as viral factories in the cell. There are many viroplasms in one infected cell, where they appear dense to electron microscopy. Very little is understood about the mechanism of viroplasm formation.

APOBEC3G is a human enzyme encoded by the APOBEC3G gene that belongs to the APOBEC superfamily of proteins. This family of proteins has been suggested to play an important role in innate anti-viral immunity. APOBEC3G belongs to the family of cytidine deaminases that catalyze the deamination of cytidine to uridine in the single stranded DNA substrate. The C-terminal domain of A3G renders catalytic activity, several NMR and crystal structures explain the substrate specificity and catalytic activity

Importin subunit alpha-3 is a protein that in humans is encoded by the KPNA3 gene.

Importin subunit alpha-7 is a protein that in humans is encoded by the KPNA6 gene.

karyopherin subunit alpha-4, also known as importin alpha-3, is a protein that in humans is encoded by the KPNA4 gene.

COP9 signalosome complex subunit 6 is a protein that in humans is encoded by the COPS6 gene.

Cullin-4A is a protein that in humans is encoded by the CUL4A gene. CUL4A belongs to the cullin family of ubiquitin ligase proteins and is highly homologous to the CUL4B protein. CUL4A regulates numerous key processes such as DNA repair, chromatin remodeling, spermatogenesis, haematopoiesis and the mitotic cell cycle. As a result, CUL4A has been implicated in several cancers and the pathogenesis of certain viruses including HIV. A component of a CUL4A complex, Cereblon, was discovered to be a major target of the teratogenic agent thalidomide.

G2/mitotic-specific cyclin-B2 is a protein that in humans is encoded by the CCNB2 gene.

Tetherin, also known as bone marrow stromal antigen 2, is a lipid raft associated protein that in humans is encoded by the BST2 gene. In addition, tetherin has been designated as CD317. This protein is constitutively expressed in mature B cells, plasma cells and plasmacytoid dendritic cells, and in many other cells, it is only expressed as a response to stimuli from IFN pathway.
The pre-integration complex (PIC) is a nucleoprotein complex of viral genetic material and associated viral and host proteins which is capable of inserting a viral genome into a host genome. The PIC forms after uncoating of a viral particle after entry into the host cell. In the case of the human immunodeficiency virus (HIV), the PIC forms after the Reverse Transcription Complex (RTC) has reverse transcribed the viral RNA into DNA. The PIC consists of viral proteins, host proteins and the viral DNA. The PIC enters the cellular nucleus through the nuclear pore complex without disrupting the nuclear envelope, thus allowing HIV and related retroviruses to replicate in non-dividing cells. Following nuclear entry, the PIC's DNA payload may be integrated into the host DNA as a "provirus".

SAM domain and HD domain-containing protein 1 is a protein that in humans is encoded by the SAMHD1 gene. SAMHD1 is a cellular enzyme, responsible for blocking replication of HIV in dendritic cells, macrophages, monocytes and resting CD4+ T lymphocytes. It is an enzyme that exhibits phosphohydrolase activity, converting deoxynucleoside triphosphates (dNTPs) to inorganic phosphate (iPPP) and a 2'-deoxynucleoside. In doing so, SAMHD1 depletes the pool of dNTPs available to a reverse transcriptase for viral cDNA synthesis and thus prevents viral replication. SAMHD1 has also shown nuclease activity. Although a ribonuclease activity was described to be required for HIV-1 restriction, recent data confirmed that SAMHD1-mediated HIV-1 restriction in cells does not involve ribonuclease activity.

Vpu is an accessory protein that in HIV is encoded by the vpu gene. Vpu stands for "Viral Protein U". The Vpu protein acts in the degradation of CD4 in the endoplasmic reticulum and in the enhancement of virion release from the plasma membrane of infected cells. Vpu induces the degradation of the CD4 viral receptor and therefore participates in the general downregulation of CD4 expression during the course of HIV infection. Vpu-mediated CD4 degradation is thought to prevent CD4-Env binding in the endoplasmic reticulum in order to facilitate proper Env assembly into virions. It is found in the membranes of infected cells, but not the virus particles themselves.

Nef is a small 27-35 kDa myristoylated protein encoded by primate lentiviruses. These include Human Immunodeficiency Viruses and Simian Immunodeficiency Virus (SIV). Nef localizes primarily to the cytoplasm but also partially to the Plasma Membrane (PM) and is one of many pathogen-expressed proteins, known as virulence factors, which function to manipulate the host's cellular machinery and thus allow infection, survival or replication of the pathogen. Nef stands for "Negative Factor" and although it is often considered dispensable for HIV-1 replication, in infected hosts the viral protein markedly elevates viral titers.

Vpr is a Human immunodeficiency virus gene and protein product.
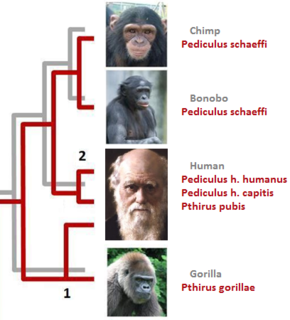
In parasitology and epidemiology, a host switch is an evolutionary change of the host specificity of a parasite or pathogen. For example, the human immunodeficiency virus used to infect and circulate in non-human primates in West-central Africa, but switched to humans in the early 20th century.
References
- ↑ Vicenzi, E.; Poli, G. (2013). "Novel factors interfering with human immunodeficiency virus-type 1 replication in vivo and in vitro". Tissue Antigens. 81 (2): 61–71. doi:10.1111/tan.12047. PMID 23330719.
- 1 2 3 4 5 Fujita, Mikako; Nomaguchi, Masako; Adachi, Akio; Otsuka, Masami (2012). "SAMHD1-Dependent and -Independent Functions of HIV-2/SIV Vpx Protein". Frontiers in Microbiology. 3: 297. doi:10.3389/fmicb.2012.00297. PMC 3415948 . PMID 22908011.
- 1 2 Laguette, Nadine; Rahm, Nadia; Sobhian, Bijan; Chable-Bessia, Christine; Münch, Jan; Snoeck, Joke; Sauter, Daniel; Switzer, William M.; et al. (2012). "Evolutionary and Functional Analyses of the Interaction between the Myeloid Restriction Factor SAMHD1 and the Lentiviral Vpx Protein". Cell Host & Microbe. 11 (2): 205–17. doi:10.1016/j.chom.2012.01.007. PMC 3595996 . PMID 22305291.
- ↑ Lahouassa, Hichem; Daddacha, Waaqo; Hofmann, Henning; Ayinde, Diana; Logue, Eric C; Dragin, Loïc; Bloch, Nicolin; Maudet, Claire; et al. (2012). "SAMHD1 restricts the replication of human immunodeficiency virus type 1 by depleting the intracellular pool of deoxynucleoside triphosphates". Nature Immunology. 13 (3): 223–8. doi:10.1038/ni.2236. PMC 3771401 . PMID 22327569.
- ↑ Wu, Xiaoyun; Conway, Joan A.; Kim, Justin; Kappes, John C. (1994). "Localization of the Vpx packaging signal within the C terminus of the human immunodeficiency virus type 2 Gag precursor protein". Journal of Virology. 68 (10): 6161–9. PMC 237035 . PMID 8083957.
- ↑ Fujita, Mikako; Otsuka, Masami; Miyoshi, Masami; Khamsri, Boonruang; Nomaguchi, Masako; Adachi, Akio (2008). "Vpx is Critical for Reverse Transcription of the Human Immunodeficiency Virus Type 2 Genome in Macrophages". Journal of Virology. 82 (15): 7752–6. doi:10.1128/JVI.01003-07. PMC 2493314 . PMID 18495778.
- 1 2 Bouzar, Amel Baya; Guiguen, François; Morin, Thierry; Villet, Stephanie; Fornazero, Claudie; Garnier, C.Éline; Gallay, Kathy; Gounel, Françoise; et al. (2003). "Specific G2 arrest of caprine cells infected with a caprine arthritis encephalitis virus expressing vpr and vpx genes from simian immunodeficiency virus". Virology. 309 (1): 41–52. doi:10.1016/S0042-6822(03)00014-X. PMID 12726725.