A provirus is a virus genome that is integrated into the DNA of a host cell. In the case of bacterial viruses (bacteriophages), proviruses are often referred to as prophages. However, proviruses are distinctly different from prophages and these terms should not be used interchangeably. Unlike prophages, proviruses do not excise themselves from the host genome when the host cell is stressed.

A retrovirus is a type of virus that inserts a DNA copy of its RNA genome into the DNA of a host cell that it invades, thus changing the genome of that cell. After invading a host cell's cytoplasm, the virus uses its own reverse transcriptase enzyme to produce DNA from its RNA genome, the reverse of the usual pattern, thus retro (backwards). The new DNA is then incorporated into the host cell genome by an integrase enzyme, at which point the retroviral DNA is referred to as a provirus. The host cell then treats the viral DNA as part of its own genome, transcribing and translating the viral genes along with the cell's own genes, producing the proteins required to assemble new copies of the virus. Many retroviruses cause serious diseases in humans, other mammals, and birds.

A reverse transcriptase (RT) is an enzyme used to generate complementary DNA (cDNA) from an RNA template, a process termed reverse transcription. Reverse transcriptases are used by viruses such as HIV and hepatitis B to replicate their genomes, by retrotransposon mobile genetic elements to proliferate within the host genome, and by eukaryotic cells to extend the telomeres at the ends of their linear chromosomes. Contrary to a widely held belief, the process does not violate the flows of genetic information as described by the classical central dogma, as transfers of information from RNA to DNA are explicitly held possible.

Retroviral integrase (IN) is an enzyme produced by a retrovirus that integrates its genetic information into that of the host cell it infects. Retroviral INs are not to be confused with phage integrases (recombinases) used in biotechnology, such as λ phage integrase, as discussed in site-specific recombination.

Retrotransposons are a type of genetic component that copy and paste themselves into different genomic locations (transposon) by converting RNA back into DNA through the reverse transcription process using an RNA transposition intermediate.
Mouse mammary tumor virus (MMTV) is a milk-transmitted retrovirus like the HTL viruses, HI viruses, and BLV. It belongs to the genus Betaretrovirus. MMTV was formerly known as Bittner virus, and previously the "milk factor", referring to the extra-chromosomal vertical transmission of murine breast cancer by adoptive nursing, demonstrated in 1936, by John Joseph Bittner while working at the Jackson Laboratory in Bar Harbor, Maine. Bittner established the theory that a cancerous agent, or "milk factor", could be transmitted by cancerous mothers to young mice from a virus in their mother's milk. The majority of mammary tumors in mice are caused by mouse mammary tumor virus.
Lentivirus is a genus of retroviruses that cause chronic and deadly diseases characterized by long incubation periods, in humans and other mammalian species. The genus includes the human immunodeficiency virus (HIV), which causes AIDS. Lentiviruses are distributed worldwide, and are known to be hosted in apes, cows, goats, horses, cats, and sheep as well as several other mammals.

Gammaretrovirus is a genus in the Retroviridae family. Example species are the murine leukemia virus and the feline leukemia virus. They cause various sarcomas, leukemias and immune deficiencies in mammals, reptiles and birds.

Endogenous retroviruses (ERVs) are endogenous viral elements in the genome that closely resemble and can be derived from retroviruses. They are abundant in the genomes of jawed vertebrates, and they comprise up to 5–8% of the human genome.
The genome and proteins of HIV (human immunodeficiency virus) have been the subject of extensive research since the discovery of the virus in 1983. "In the search for the causative agent, it was initially believed that the virus was a form of the Human T-cell leukemia virus (HTLV), which was known at the time to affect the human immune system and cause certain leukemias. However, researchers at the Pasteur Institute in Paris isolated a previously unknown and genetically distinct retrovirus in patients with AIDS which was later named HIV." Each virion comprises a viral envelope and associated matrix enclosing a capsid, which itself encloses two copies of the single-stranded RNA genome and several enzymes. The discovery of the virus itself occurred two years following the report of the first major cases of AIDS-associated illnesses.
The murine leukemia viruses are retroviruses named for their ability to cause cancer in murine (mouse) hosts. Some MLVs may infect other vertebrates. MLVs include both exogenous and endogenous viruses. Replicating MLVs have a positive sense, single-stranded RNA (ssRNA) genome that replicates through a DNA intermediate via the process of reverse transcription.
Human foamy virus (HFV) is a retrovirus and specifically belongs to the genus Spumavirus. The spumaviruses are complex and significantly different from the other six genera of retroviruses in several ways. The foamy viruses derive their name from the characteristic ‘foamy’ appearance of the cytopathic effect (CPE) induced in the cells. Foamy virus in humans occurs only as a result of zoonotic infection.

APOBEC3G is a human enzyme encoded by the APOBEC3G gene that belongs to the APOBEC superfamily of proteins. This family of proteins has been suggested to play an important role in innate anti-viral immunity. APOBEC3G belongs to the family of cytidine deaminases that catalyze the deamination of cytidine to uridine in the single stranded DNA substrate. The C-terminal domain of A3G renders catalytic activity, several NMR and crystal structures explain the substrate specificity and catalytic activity.
Simian foamy virus (SFV) is a species of the genus Spumavirus that belongs to the family of Retroviridae. It has been identified in a wide variety of primates, including prosimians, New World and Old World monkeys, as well as apes, and each species has been shown to harbor a unique (species-specific) strain of SFV, including African green monkeys, baboons, macaques, and chimpanzees. As it is related to the more well-known retrovirus human immunodeficiency virus (HIV), its discovery in primates has led to some speculation that HIV may have been spread to the human species in Africa through contact with blood from apes, monkeys, and other primates, most likely through bushmeat-hunting practices.

The retroviral psi packaging element, also known as the Ψ RNA packaging signal, is a cis-acting RNA element identified in the genomes of the retroviruses Human immunodeficiency virus (HIV) and Simian immunodeficiency virus (SIV). It is involved in regulating the essential process of packaging the retroviral RNA genome into the viral capsid during replication. The final virion contains a dimer of two identical unspliced copies of the viral genome.
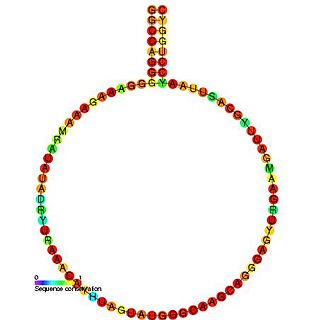
HIV gag stem loop 3 (GSL3) is a secondary structural component of the Retroviral Psi packaging element, also known as the psi recognition element. This domain plays a major role in RNA packaging and is located the 5’ untranslated region of the unspliced HIV-1 genome. GSL3 is known to direct specific packaging of HIV-1 genomic RNA. While deletion of GSL3 leads to decreases in both viral RNA packaging and dimerization, mutagenic studies have shown that it does not eliminate encapsulation of retroviral RNA.

LTR retrotransposons are class I transposable element characterized by the presence of long terminal repeats (LTRs) directly flanking an internal coding region. As retrotransposons, they mobilize through reverse transcription of their mRNA and integration of the newly created cDNA into another location. Their mechanism of retrotransposition is shared with retroviruses, with the difference that most LTR-retrotransposons do not form infectious particles that leave the cells and therefore only replicate inside their genome of origin. Those that do (occasionally) form virus-like particles are classified under Ortervirales.
Bovine immunodeficiency virus (BIV) is a retrovirus belonging to the genus Lentivirus. It is similar to the human immunodeficiency virus (HIV) and infects cattle. The cells primarily infected are lymphocytes and monocytes/macrophages.
Mason-Pfizer monkey virus (M-PMV), formerly Simian retrovirus (SRV), is a species of retroviruses that usually infect and cause a fatal immune deficiency in Asian macaques. The ssRNA virus appears sporadically in mammary carcinoma of captive macaques at breeding facilities which expected as the natural host, but the prevalence of this virus in feral macaques remains unknown. M-PMV was transmitted naturally by virus-containing body fluids, via biting, scratching, grooming, and fighting. Cross contaminated instruments or equipment (fomite) can also spread this virus among animals.
Feline foamy virus or Feline syncytial virus is a retrovirus and belongs to the family Retroviridae and the subfamily Spumaretrovirinae. It shares the genus Felispumavirus with only Puma feline foamy virus. There has been controversy on whether FeFV is nonpathogenic as the virus is generally asymptomatic in affected cats and does not cause disease. However, some changes in kidney and lung tissue have been observed over time in cats affected with FeFV, which may or may not be directly affiliated. This virus is fairly common and infection rates gradually increase with a cat's age. Study results from antibody examinations and PCR analysis have shown that over 70% of felines over 9 years old were seropositive for Feline foamy virus. Viral infections are similar between male and female domesticated cats whereas in the wild, more feral females cats are affected with FeFV.










