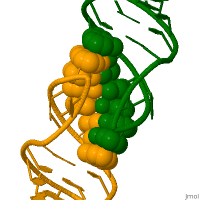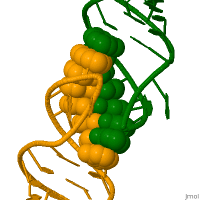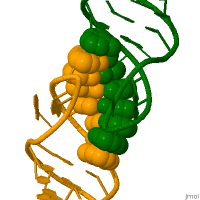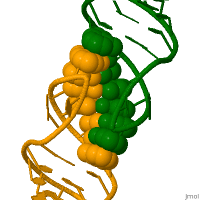
RNA molecules perform their function in living cells by adopting specific and highly complex 3-dimensional structures. It is believed that recombination may be initiated by the kissing loops. Recombination is critical to successful evolution, especially in the adaptation and survival of viruses. [6]
In retroviruses
Retroviruses are viruses that silently replicate inside a host, keeping it alive until the replication is completed and the host is no longer needed. The genomic RNA of retroviruses is linked non-covalently to the dimer linkage structure (DLS), a non-coding region in the 5' UTR. For the kissing loop interaction to occur, there is a triple interaction that involves a 5'-flanking purine and 2 centralized bases in the complementary strand. This interaction transcripts in the major groove of the kissing loop dimer. [7]
The human immunodeficiency virus (HIV) is a retrovirus that can be transmitted via the interactions of bodily fluids. There are two types of HIV viruses, human immunodeficiency virus type 1 (HIV-1) and human immunodeficiency virus type 2 (HIV-2). From the two types of the Human Immunodeficiency Virus strain, HIV-1 is the most common strain. The RNA of the HIV-1 virus uses the kissing stem loop interaction as a means of recognition, the main step of interaction of the dimerization initiation site (DIS) to form the duplex. To study the kissing stem-loop loop interaction, It was seen that the Dimerization initiation site (DIS) complex was essential to the replication of the HIV type 1 virus in the eukaryotic cell, and any changes to the stem loop structure diminished the dimerization interaction. Experimentally, it has been seen that, in vivo, mutating the Dimerization initiation site (DIS) obstructed the dimerization of the DIS complex. [8]