
In molecular biology, molecular chaperones are proteins that assist the conformational folding or unfolding of large proteins or macromolecular protein complexes. There are a number of classes of molecular chaperones, all of which function to assist large proteins in proper protein folding during or after synthesis, and after partial denaturation. Chaperones are also involved in the translocation of proteins for proteolysis.
The 70 kilodalton heat shock proteins are a family of conserved ubiquitously expressed heat shock proteins. Proteins with similar structure exist in virtually all living organisms. Intracellularly localized Hsp70s are an important part of the cell's machinery for protein folding, performing chaperoning functions, and helping to protect cells from the adverse effects of physiological stresses. Additionally, membrane-bound Hsp70s have been identified as a potential target for cancer therapies and their extracellularly localized counterparts have been identified as having both membrane-bound and membrane-free structures.

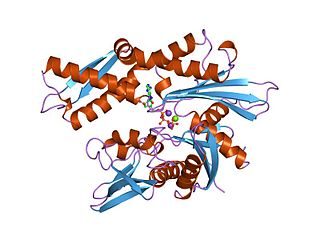
Hsp90 is a chaperone protein that assists other proteins to fold properly, stabilizes proteins against heat stress, and aids in protein degradation. It also stabilizes a number of proteins required for tumor growth, which is why Hsp90 inhibitors are investigated as anti-cancer drugs.

The nucleoid is an irregularly shaped region within the prokaryotic cell that contains all or most of the genetic material. The chromosome of a prokaryote is circular, and its length is very large compared to the cell dimensions, so it needs to be compacted in order to fit. In contrast to the nucleus of a eukaryotic cell, it is not surrounded by a nuclear membrane. Instead, the nucleoid forms by condensation and functional arrangement with the help of chromosomal architectural proteins and RNA molecules as well as DNA supercoiling. The length of a genome widely varies and a cell may contain multiple copies of it.
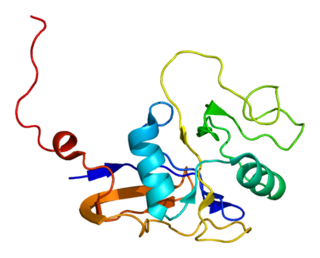
GroEL is a protein which belongs to the chaperonin family of molecular chaperones, and is found in many bacteria. It is required for the proper folding of many proteins. To function properly, GroEL requires the lid-like cochaperonin protein complex GroES. In eukaryotes the organellar proteins Hsp60 and Hsp10 are structurally and functionally nearly identical to GroEL and GroES, respectively, due to their endosymbiotic origin.
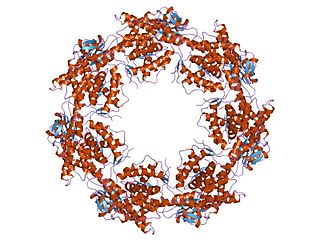
HSP60, also known as chaperonins (Cpn), is a family of heat shock proteins originally sorted by their 60kDa molecular mass. They prevent misfolding of proteins during stressful situations such as high heat, by assisting protein folding. HSP60 belong to a large class of molecules that assist protein folding, called molecular chaperones.
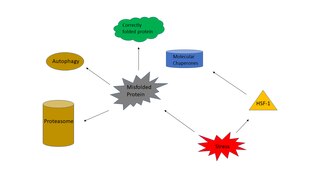
The heat shock response (HSR) is a cell stress response that increases the number of molecular chaperones to combat the negative effects on proteins caused by stressors such as increased temperatures, oxidative stress, and heavy metals. In a normal cell, proteostasis must be maintained because proteins are the main functional units of the cell. Many proteins take on a defined configuration in a process known as protein folding in order to perform their biological functions. If these structures are altered, critical processes could be affected, leading to cell damage or death. The heat shock response can be employed under stress to induce the expression of heat shock proteins (HSP), many of which are molecular chaperones, that help prevent or reverse protein misfolding and provide an environment for proper folding.

The heat shock proteins HslV and HslU are expressed in many bacteria such as E. coli in response to cell stress. The hslV protein is a protease and the hslU protein is an ATPase; the two form a symmetric assembly of four stacked rings, consisting of an hslV dodecamer bound to an hslU hexamer, with a central pore in which the protease and ATPase active sites reside. The hslV protein degrades unneeded or damaged proteins only when in complex with the hslU protein in the ATP-bound state. HslV is thought to resemble the hypothetical ancestor of the proteasome, a large protein complex specialized for regulated degradation of unneeded proteins in eukaryotes, many archaea, and a few bacteria. HslV bears high similarity to core subunits of proteasomes.

Heat shock 70 kDa protein 1, also termed Hsp72, is a protein that in humans is encoded by the HSPA1A gene. As a member of the heat shock protein 70 family and a chaperone protein, it facilitates the proper folding of newly translated and misfolded proteins, as well as stabilize or degrade mutant proteins. In addition, Hsp72 also facilitates DNA repair. Its functions contribute to biological processes including signal transduction, apoptosis, protein homeostasis, and cell growth and differentiation. It has been associated with an extensive number of cancers, neurodegenerative diseases, cell senescence and aging, and inflammatory diseases such as Diabetes mellitus type 2 and rheumatoid arthritis.

Heat shock factor 1 (HSF1) is a protein that in humans is encoded by the HSF1 gene. HSF1 is highly conserved in eukaryotes and is the primary mediator of transcriptional responses to proteotoxic stress with important roles in non-stress regulation such as development and metabolism.

Heat shock protein HSP 90-beta also called HSP90beta is a protein that in humans is encoded by the HSP90AB1 gene.

Binding immunoglobulin protein (BiP) also known as 78 kDa glucose-regulated protein (GRP-78) or heat shock 70 kDa protein 5 (HSPA5) is a protein that in humans is encoded by the HSPA5 gene.

FourU thermometers are a class of non-coding RNA thermometers found in Salmonella. They are named 'FourU' due to the four highly conserved uridine nucleotides found directly opposite the Shine-Dalgarno sequence on hairpin II (pictured). RNA thermometers such as FourU control regulation of temperature via heat shock proteins in many prokaryotes. FourU thermometers are relatively small RNA molecules, only 57 nucleotides in length, and have a simple two-hairpin structure.

In molecular biology, chaperone DnaJ, also known as Hsp40, is a molecular chaperone protein. It is expressed in a wide variety of organisms from bacteria to humans.

An RNA thermometer is a temperature-sensitive non-coding RNA molecule which regulates gene expression. RNA thermometers often regulate genes required during either a heat shock or cold shock response, but have been implicated in other regulatory roles such as in pathogenicity and starvation.
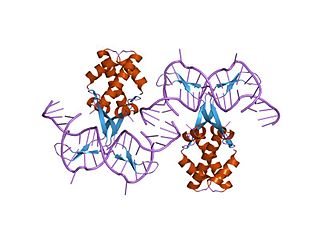
In molecular biology, bacterial DNA binding proteins are a family of small, usually basic proteins of about 90 residues that bind DNA and are known as histone-like proteins. Since bacterial binding proteins have a diversity of functions, it has been difficult to develop a common function for all of them. They are commonly referred to as histone-like and have many similar traits with the eukaryotic histone proteins. Eukaryotic histones package DNA to help it to fit in the nucleus, and they are known to be the most conserved proteins in nature. Examples include the HU protein in Escherichia coli, a dimer of closely related alpha and beta chains and in other bacteria can be a dimer of identical chains. HU-type proteins have been found in a variety of bacteria and archaea, and are also encoded in the chloroplast genome of some algae. The integration host factor (IHF), a dimer of closely related chains which is suggested to function in genetic recombination as well as in translational and transcriptional control is found in Enterobacteria and viral proteins including the African swine fever virus protein A104R.
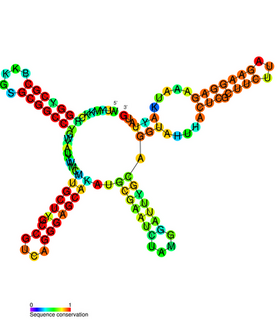
The IbpB thermometer is an RNA thermometer element found in the ibpAB operon. The operon contains two heat-shock genes, encoding inclusion body binding proteins A and B (IbpA/B), and is the most drastically upregulated operon under heat-shock in Escherichia coli.

The universal stress protein (USP) domain is a superfamily of conserved genes which can be found in bacteria, archaea, fungi, protozoa and plants. Proteins containing the domain are induced by many environmental stressors such as nutrient starvation, drought, extreme temperatures, high salinity, and the presence of uncouplers, antibiotics and metals.

Sue Hengren Wickner is an American biochemist and geneticist who is a distinguished investigator and the head of the DNA Molecular Biology section of the National Institutes of Health. Her laboratory is under the National Cancer Institute and is located in the Center for Cancer Research (NCI/CCR).

GrpE is a bacterial nucleotide exchange factor that is important for regulation of protein folding machinery, as well as the heat shock response. It is a heat-inducible protein and during stress it prevents unfolded proteins from accumulating in the cytoplasm. Accumulation of unfolded proteins in the cytoplasm can lead to cell death.



















