
Ribosomes are macromolecular machines, found within all living cells, that perform biological protein synthesis. Ribosomes link amino acids together in the order specified by the codons of messenger RNA (mRNA) molecules to form polypeptide chains. Ribosomes consist of two major components: the small and large ribosomal subunits. Each subunit consists of one or more ribosomal RNA (rRNA) molecules and many ribosomal proteins. The ribosomes and associated molecules are also known as the translational apparatus.

Microbial ecology is the ecology of microorganisms: their relationship with one another and with their environment. It concerns the three major domains of life—Eukaryota, Archaea, and Bacteria—as well as viruses.

Metagenomics is the study of genetic material recovered directly from environmental samples. The broad field may also be referred to as environmental genomics, ecogenomics or community genomics.

16S ribosomal RNA is the RNA component of the 30S subunit of a prokaryotic ribosome. It binds to the Shine-Dalgarno sequence and provides most of the SSU structure.

Microbiota are "ecological communities of commensal, symbiotic and pathogenic microorganisms" found in and on all multicellular organisms studied to date from plants to animals. Microbiota include bacteria, archaea, protists, fungi and viruses. Microbiota have been found to be crucial for immunologic, hormonal and metabolic homeostasis of their host. The term microbiome describes either the collective genomes of the microorganisms that reside in an environmental niche or the microorganisms themselves.
Giant, Ornate, Lake- and Lactobacillales-Derived (GOLLD) RNA is a conserved RNA structure present in bacteria. GOLLD RNAs were originally detected based on metagenome sequences of DNA isolated from Lake Gatun in Panama. However, they are known to be present in at least eight strains of cultivated bacteria. GOLLD RNAs are extraordinarily large compared to other RNAs with a conserved, complex secondary structure, and average roughly 800 nucleotides. Such large, complex RNAs are often ribozymes, although the biochemical function of GOLLD RNAs remains unknown. The discovery of large RNAs like GOLLD RNAs among bacteria that are mostly uncultivated under laboratory conditions suggests that many other unusually large RNAs might be found in bacteria that have not yet been studied.
HNH Endonuclease-Associated RNA and ORF (HEARO) RNAs conform to a conserved RNA structure that was identified in bacteria by bioinformatics. HEARO RNAs average roughly 300 nucleotides, which is comparable to the size of many ribozymes, which catalyze chemical reactions.

A wide variety of non-coding RNAs have been identified in various species of organisms known to science. However, RNAs have also been identified in "metagenomics" sequences derived from samples of DNA or RNA extracted from the environment, which contain unknown species. Initial work in this area detected homologs of known bacterial RNAs in such metagenome samples. Many of these RNA sequences were distinct from sequences within cultivated bacteria, and provide the potential for additional information on the RNA classes to which they belong.

The IMES-2 RNA motif is a conserved RNA structure that was identified by a study based on metagenomics and bioinformatics, and the underlying RNA sequences were identified independently by a similar earlier study. These RNAs are present in environmental sequences, and when discovered were not known to be present in any cultivated species. However, an IMES-2 RNA has been detected in alphaproteobacterium HIMB114, which is classified in the SAR11 clade of marine bacteria. This finding fits with earlier predictions that species that use IMES-2 RNAs are most closely related to alphaproteobacteria. IMES-2 RNAs are exceptionally abundant, as twice as many IMES-2 RNAs were found as ribosomes in RNAs sampled from the Pacific Ocean. Only two bacterial RNAs are known to be more highly transcribed than ribosomes.

The IMES-3 RNA motif is a conserved RNA structure that was identified based on metagenomics and bioinformatics, and the underlying RNA sequences were identified independently by an earlier study. These RNAs are present in environmental sequences, and as of 2009 are not known to be present in any cultivated species. IMES-3 RNAs are abundant in comparison to ribosomes in RNAs sampled from the Pacific Ocean.

The IMES-4 RNA motif is a conserved RNA structure that was identified in marine environmental sequences by metagenomics and bioinformatics. These RNAs are present in environmental sequences, and as of 2009 are not known to be present in any cultivated species. IMES-4 RNAs are fairly abundant in comparison to ribosomes in RNAs sampled from the Pacific Ocean.

The Dictyoglomi-1 RNA motif is a conserved RNA structure that was discovered via bioinformatics. Only four instances of the RNA were detected, and all are in the bacterial phylum Dictyoglomi, whose members have not been extensively studied. The RNA might have a cis-regulatory role, but the evidence is ambiguous. Because of the few instances of Dictyoglomi-1 RNAs known, it is also unknown whether the RNA structure might extend further in the 5′ or 3′ direction, or in both directions.

The Gut-1 RNA motif is a conserved RNA structure identified by bioinformatics. These RNAs are present in environmental sequences, and as of 2010 are not known to be present in any species that has been grown under laboratory conditions. Gut-1 RNA is exclusively found in DNA from uncultivated bacteria present in samples from the human gut.
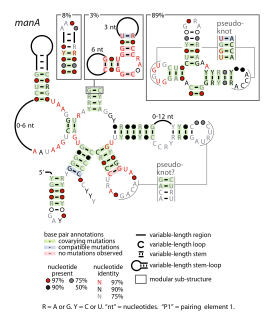
The manA RNA motif refers to a conserved RNA structure that was identified by bioinformatics. Instances of the manA RNA motif were detected in bacteria in the genus Photobacterium and phages that infect certain kinds of cyanobacteria. However, most predicted manA RNA sequences are derived from DNA collected from uncultivated marine bacteria. Almost all manA RNAs are positioned such that they might be in the 5' untranslated regions of protein-coding genes, and therefore it was hypothesized that manA RNAs function as cis-regulatory elements. Given the relative complexity of their secondary structure, and their hypothesized cis-regulatory role, they might be riboswitches.

The Termite-flg RNA motif is a conserved RNA structure identified by bioinformatics. Genomic sequences corresponding to Termite-flg RNAs have been identified only in uncultivated bacteria present in the termite hindgut. As of 2010 it has not been identified in the DNA of any cultivated species, and is thus an example of RNAs present in environmental samples.

The Whalefall-1 RNA motif refers to a conserved RNA structure that was discovered using bioinformatics. Structurally, the motif consists of two stem-loops, the second of which is often terminated by a CUUG tetraloop, which is an energetically favorable RNA sequence. Whalefall-1 RNAs are found only in DNA extracted from uncultivated bacteria found on whale fall, i.e., a whale carcass. As of 2010, Whalefall-1 RNAs have not been detected in any known, cultivated species of bacteria, and are thus one of several RNAs present in environmental samples.

Bacterial phyla constitute the major lineages of the domain Bacteria. While the exact definition of a bacterial phylum is debated, a popular definition is that a bacterial phylum is a monophyletic lineage of bacteria whose 16S rRNA genes share a pairwise sequence identity of ~75% or less with those of the members of other bacterial phyla.
PICRUSt is a bioinformatics software package. The name is an abbreviation for Phylogenetic Investigation of Communities by Reconstruction of Unobserved States.

The Rumen-Originating, Ornate, Large (ROOL) RNA motif was originally discovered by bioinformatics by analyzing metagenomic sequences from cow rumen. ROOL RNAs are found in a variety of bacterial species and apparently do not code for proteins. The RNA has a complex RNA secondary structure and its average size of 581 nucleotides is unusually large for bacterial non-coding RNAs. This large size and structural complexity for a bacterial RNA is consistent with properties of large ribozymes.

The Ocean-VII RNA motif is a conserved RNA structure that was discovered by bioinformatics. Ocean-VII motifs are found in metagenomic sequences isolated from various marine environments, and are not yet known in any classified organism. This environmental context is similar to other marine RNAs that were found previously by predominantly bioinformatic or experimental methods.

















