Related Research Articles
In biology and biochemistry, protease inhibitors, or antiproteases, are molecules that inhibit the function of proteases. Many naturally occurring protease inhibitors are proteins.
Ferredoxins are iron–sulfur proteins that mediate electron transfer in a range of metabolic reactions. The term "ferredoxin" was coined by D.C. Wharton of the DuPont Co. and applied to the "iron protein" first purified in 1962 by Mortenson, Valentine, and Carnahan from the anaerobic bacterium Clostridium pasteurianum.
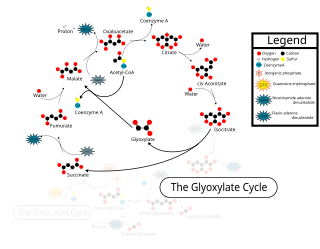
The glyoxylate cycle, a variation of the tricarboxylic acid cycle, is an anabolic pathway occurring in plants, bacteria, protists, and fungi. The glyoxylate cycle centers on the conversion of acetyl-CoA to succinate for the synthesis of carbohydrates. In microorganisms, the glyoxylate cycle allows cells to utilize two carbons, such as acetate, to satisfy cellular carbon requirements when simple sugars such as glucose or fructose are not available. The cycle is generally assumed to be absent in animals, with the exception of nematodes at the early stages of embryogenesis. In recent years, however, the detection of malate synthase (MS) and isocitrate lyase (ICL), key enzymes involved in the glyoxylate cycle, in some animal tissue has raised questions regarding the evolutionary relationship of enzymes in bacteria and animals and suggests that animals encode alternative enzymes of the cycle that differ in function from known MS and ICL in non-metazoan species.
Oxaloacetate decarboxylase is a carboxy-lyase involved in the conversion of oxaloacetate into pyruvate.
In enzymology, a holocytochrome-c synthase is an enzyme that catalyzes the chemical reaction

Isocitrate lyase, or ICL, is an enzyme in the glyoxylate cycle that catalyzes the cleavage of isocitrate to succinate and glyoxylate. Together with malate synthase, it bypasses the two decarboxylation steps of the tricarboxylic acid cycle and is used by bacteria, fungi, and plants.

In enzymology, a methylisocitrate lyase is an enzyme that catalyzes the chemical reaction
In enzymology, a malate synthase (EC 2.3.3.9) is an enzyme that catalyzes the chemical reaction
The class C G-protein-coupled receptors are a class of G-protein coupled receptors that include the metabotropic glutamate receptors and several additional receptors.
In enzymology, a carboxyvinyl-carboxyphosphonate phosphorylmutase (EC 2.7.8.23) is an enzyme that catalyzes the chemical reaction
Plant defensins are a family of small, cysteine-rich proteins found in plants that serve to defend them against parasites.
Aminotransferase class-V is an evolutionary conserved protein domain. This domain is found in amino transferases, and other enzymes including cysteine desulphurase EC:4.4.1.-.
A number of enzymes, belonging to the lyase class, for which fumarate is a substrate, have been shown to share a short conserved sequence around a methionine which is probably involved in the catalytic activity of this type of enzymes. The following are examples of members of this family:

In molecular biology, a carbohydrate-binding module (CBM) is a protein domain found in carbohydrate-active enzymes. The majority of these domains have carbohydrate-binding activity. Some of these domains are found on cellulosomal scaffoldin proteins. CBMs were previously known as cellulose-binding domains. CBMs are classified into numerous families, based on amino acid sequence similarity. There are currently 64 families of CBM in the CAZy database.

The SET domain is a protein domain. It was originally identified as part of a larger conserved region present in the Drosophila Trithorax protein and was subsequently identified in the Drosophila Su(var)3-9 and 'Enhancer of zeste' proteins, from which the acronym SET is derived [Su(var)3-9, Enhancer-of-zeste and Trithorax].

In molecular biology, Glycoside hydrolase family 2 is a family of glycoside hydrolases.

In molecular biology, glycoside hydrolase family 56 is a family of glycoside hydrolases.

In molecular biology, the H2TH domain is a DNA-binding domain found in DNA glycosylase/AP lyase enzymes, which are involved in base excision repair of DNA damaged by oxidation or by mutagenic agents. Most damage to bases in DNA is repaired by the base excision repair pathway. These enzymes are primarily from bacteria, and have both DNA glycosylase activity EC 3.2.2.- and AP lyase activity EC 4.2.99.18. Examples include formamidopyrimidine-DNA glycosylases and endonuclease VIII (Nei).

In molecular biology, the isocitrate/isopropylmalate dehydrogenase family is a protein family consisting of the evolutionary related enzymes isocitrate dehydrogenase, 3-isopropylmalate dehydrogenase and tartrate dehydrogenase.

4-Hydroxy-tetrahydrodipicolinate synthase (EC 4.3.3.7, dihydrodipicolinate synthase, dihydropicolinate synthetase, dihydrodipicolinic acid synthase, L-aspartate-4-semialdehyde hydro-lyase (adding pyruvate and cyclizing), dapA (gene)) is an enzyme with the systematic name L-aspartate-4-semialdehyde hydro-lyase (adding pyruvate and cyclizing; (4S)-4-hydroxy-2,3,4,5-tetrahydro-(2S)-dipicolinate-forming). This enzyme catalyses the following chemical reaction
References
- ↑ Beeching JR (1989). "High sequence conservation between isocitrate lyase from Escherichia coli and Ricinus communis". Protein Seq. Data Anal. 2 (6): 463–466. PMID 2696959.
- ↑ Tanaka A, Atomi H, Ueda M, Hikida M, Hishida T, Teranishi Y (1990). "Peroxisomal isocitrate lyase of the n-alkane-assimilating yeast Candida tropicalis: gene analysis and characterization". J. Biochem. 107 (2): 262–266. doi:10.1093/oxfordjournals.jbchem.a123036. PMID 2361956.
| This protein-related article is a stub. You can help Wikipedia by expanding it. |