Related Research Articles
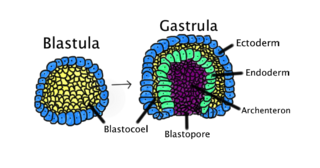
Gastrulation is the stage in the early embryonic development of most animals, during which the blastula, or in mammals the blastocyst, is reorganized into a two-layered or three-layered embryo known as the gastrula. Before gastrulation, the embryo is a continuous epithelial sheet of cells; by the end of gastrulation, the embryo has begun differentiation to establish distinct cell lineages, set up the basic axes of the body, and internalized one or more cell types including the prospective gut.

The European Molecular Biology Laboratory (EMBL) is an intergovernmental organization dedicated to molecular biology research and is supported by 29 member states, two prospect member states, and one associate member state. EMBL was created in 1974 and is funded by public research money from its member states. Research at EMBL is conducted by more than 110 independent research groups and service teams covering the spectrum of molecular biology.

The Babraham Institute is a life sciences research institution focussing on healthy ageing. The Babraham Institute is based on the Babraham Research Campus, partly occupying a former manor house, but also laboratory and science facility buildings on the campus, surrounded by an extensive parkland estate, just south of Cambridge, England. It is an independent and charitable organization which is involved in biomedical research, including healthy aging and molecular biology. The director is Dr Simon Cook who also leads the Institute's signalling research programme.

John Frederick William Birney is joint director of EMBL's European Bioinformatics Institute (EMBL-EBI), in Hinxton, Cambridgeshire and deputy director general of the European Molecular Biology Laboratory (EMBL). He also serves as non-executive director of Genomics England, chair of the Global Alliance for Genomics and Health (GA4GH) and honorary professor of bioinformatics at the University of Cambridge. Birney has made significant contributions to genomics, through his development of innovative bioinformatics and computational biology tools. He previously served as an associate faculty member at the Wellcome Trust Sanger Institute.

The Wellcome Genome Campus is a scientific research campus built in the grounds of Hinxton Hall, Hinxton in Cambridgeshire, England.

Dame Janet Maureen Thornton, is a senior scientist and director emeritus at the European Bioinformatics Institute (EBI), part of the European Molecular Biology Laboratory (EMBL). She is one of the world's leading researchers in structural bioinformatics, using computational methods to understand protein structure and function. She served as director of the EBI from October 2001 to June 2015, and played a key role in ELIXIR.
The Reference Sequence (RefSeq) database is an open access, annotated and curated collection of publicly available nucleotide sequences and their protein products. RefSeq was introduced in 2000. This database is built by National Center for Biotechnology Information (NCBI), and, unlike GenBank, provides only a single record for each natural biological molecule for major organisms ranging from viruses to bacteria to eukaryotes.

ChEMBL or ChEMBLdb is a manually curated chemical database of bioactive molecules with drug inducing properties. It is maintained by the European Bioinformatics Institute (EBI), of the European Molecular Biology Laboratory (EMBL), based at the Wellcome Trust Genome Campus, Hinxton, UK.
Edith Heard is a British-French researcher in epigenetics who has been serving as the Director General of the European Molecular Biology Laboratory (EMBL) since January 2019. She is also Professor at the Collège de France, holding the Chair of Epigenetics and Cellular Memory. In 2025 she will become CEO of the Crick Institute in London, U.K.

Dana Pe'er, Chair and Professor in Computational and Systems Biology Program at Sloan Kettering Institute is a researcher in computational systems biology. A Howard Hughes Medical Institute (HHMI) Investigator since 2021, she was previously a professor at Columbia Department of Biological Sciences. Pe'er's research focuses on understanding the organization, function and evolution of molecular networks, particularly how genetic variations alter the regulatory network and how these genetic variations can cause cancer.

Alexander George Bateman is a computational biologist and Head of Protein Sequence Resources at the European Bioinformatics Institute (EBI), part of the European Molecular Biology Laboratory (EMBL) in Cambridge, UK. He has led the development of the Pfam biological database and introduced the Rfam database of RNA families. He has also been involved in the use of Wikipedia for community-based annotation of biological databases.

Sarah Amalia Teichmann is a German scientist who is head of cellular genetics at the Wellcome Sanger Institute and a visiting research group leader at the European Bioinformatics Institute (EMBL-EBI). She serves as director of research in the Cavendish Laboratory, at the University of Cambridge and a senior research fellow at Churchill College, Cambridge.
Single-cell transcriptomics examines the gene expression level of individual cells in a given population by simultaneously measuring the RNA concentration of hundreds to thousands of genes. Single-cell transcriptomics makes it possible to unravel heterogeneous cell populations, reconstruct cellular developmental pathways, and model transcriptional dynamics — all previously masked in bulk RNA sequencing.
Transcriptomics technologies are the techniques used to study an organism's transcriptome, the sum of all of its RNA transcripts. The information content of an organism is recorded in the DNA of its genome and expressed through transcription. Here, mRNA serves as a transient intermediary molecule in the information network, whilst non-coding RNAs perform additional diverse functions. A transcriptome captures a snapshot in time of the total transcripts present in a cell. Transcriptomics technologies provide a broad account of which cellular processes are active and which are dormant. A major challenge in molecular biology is to understand how a single genome gives rise to a variety of cells. Another is how gene expression is regulated.

Trajectory inference or pseudotemporal ordering is a computational technique used in single-cell transcriptomics to determine the pattern of a dynamic process experienced by cells and then arrange cells based on their progression through the process. Single-cell protocols have much higher levels of noise than bulk RNA-seq, so a common step in a single-cell transcriptomics workflow is the clustering of cells into subgroups. Clustering can contend with this inherent variation by combining the signal from many cells, while allowing for the identification of cell types. However, some differences in gene expression between cells are the result of dynamic processes such as the cell cycle, cell differentiation, or response to an external stimuli. Trajectory inference seeks to characterize such differences by placing cells along a continuous path that represents the evolution of the process rather than dividing cells into discrete clusters. In some methods this is done by projecting cells onto an axis called pseudotime which represents the progression through the process.

Coiled-coil domain containing 60 is a protein that in humans is encoded by the CCDC60 gene that is most highly expressed in the trachea, salivary glands, bladder, cervix, and epididymis.
Julius Brennecke is a German molecular biologist and geneticist. He is a Senior Group Leader at the Institute of Molecular Biotechnology. (IMBA) of the Austrian Academy of Sciences in Vienna.

Stein Aerts is a Belgian bio-engineer and computational biologist. He leads the Laboratory of Computational Biology at VIB and KU Leuven, and is director of VIB.AI, the VIB Center for AI & Computational Biology. He has received several accolades for his research into the workings of the genomic regulatory code.

Single-cell multi-omics integration describes a suite of computational methods used to harmonize information from multiple "omes" to jointly analyze biological phenomena. This approach allows researchers to discover intricate relationships between different chemical-physical modalities by drawing associations across various molecular layers simultaneously. Multi-omics integration approaches can be categorized into four broad categories: Early integration, intermediate integration, late integration methods. Multi-omics integration can enhance experimental robustness by providing independent sources of evidence to address hypotheses, leveraging modality-specific strengths to compensate for another's weaknesses through imputation, and offering cell-type clustering and visualizations that are more aligned with reality
References
- ↑ "John Marioni". Mathematics Genealogy Project. Retrieved 2022-01-05.
- ↑ Institute, EMBL’s European Bionformatics. "John Marioni, Visitor | People | EMBL's European Bionformatics Institute". www.ebi.ac.uk. Retrieved 2024-08-12.
- ↑ "Marioni Group". Cancer Research UK Cambridge Institute. Retrieved 2022-01-05.
- ↑ "Human Cell Atlas Working Groups". www.humancellatlas.org. Retrieved 2022-01-05.
- ↑ "John Marioni publications indexed by Google Scholar". scholar.google.com. Retrieved 2022-01-05.
- ↑ Institute, EMBL’s European Bionformatics. "John Marioni, Visitor | People | EMBL's European Bionformatics Institute". www.ebi.ac.uk. Retrieved 2023-12-07.
- ↑ Cell Symposia | Speaker | John Marioni, EMBL-European Bioinformatics Institute, UK
- 1 2 "2021 United Kingdom Award Finalist". blavatnikawards.org. Retrieved 2022-01-05.