Deamination is the removal of an amino group from a molecule. Enzymes that catalyse this reaction are called deaminases.

The CpG sites or CG sites are regions of DNA where a cytosine nucleotide is followed by a guanine nucleotide in the linear sequence of bases along its 5' → 3' direction. CpG sites occur with high frequency in genomic regions called CpG islands.

A DNA polymerase is a member of a family of enzymes that catalyze the synthesis of DNA molecules from nucleoside triphosphates, the molecular precursors of DNA. These enzymes are essential for DNA replication and usually work in groups to create two identical DNA duplexes from a single original DNA duplex. During this process, DNA polymerase "reads" the existing DNA strands to create two new strands that match the existing ones. These enzymes catalyze the chemical reaction

DNA repair is a collection of processes by which a cell identifies and corrects damage to the DNA molecules that encodes its genome. In human cells, both normal metabolic activities and environmental factors such as radiation can cause DNA damage, resulting in tens of thousands of individual molecular lesions per cell per day. Many of these lesions cause structural damage to the DNA molecule and can alter or eliminate the cell's ability to transcribe the gene that the affected DNA encodes. Other lesions induce potentially harmful mutations in the cell's genome, which affect the survival of its daughter cells after it undergoes mitosis. As a consequence, the DNA repair process is constantly active as it responds to damage in the DNA structure. When normal repair processes fail, and when cellular apoptosis does not occur, irreparable DNA damage may occur, including double-strand breaks and DNA crosslinkages. This can eventually lead to malignant tumors, or cancer as per the two-hit hypothesis.

A molecular lesion or point lesion is damage to the structure of a biological molecule such as DNA, RNA, or protein. This damage may result in the reduction or absence of normal function, and in rare cases the gain of a new function. Lesions in DNA may consist of breaks or other changes in chemical structure of the helix, ultimately preventing transcription. Meanwhile, lesions in proteins consist of both broken bonds and improper folding of the amino acid chain. While many nucleic acid lesions are general across DNA and RNA, some are specific to one, such as thymine dimers being found exclusively in DNA. Several cellular repair mechanisms exist, ranging from global to specific, in order to prevent lasting damage resulting from lesions.

Activation-induced cytidine deaminase, also known as AICDA, AID and single-stranded DNA cytosine deaminase, is a 24 kDa enzyme which in humans is encoded by the AICDA gene. It creates mutations in DNA by deamination of cytosine base, which turns it into uracil. In other words, it changes a C:G base pair into a U:G mismatch. The cell's DNA replication machinery recognizes the U as a T, and hence C:G is converted to a T:A base pair. During germinal center development of B lymphocytes, AID also generates other types of mutations, such as C:G to A:T. The mechanism by which these other mutations are created is not well understood. It is a member of the APOBEC family.
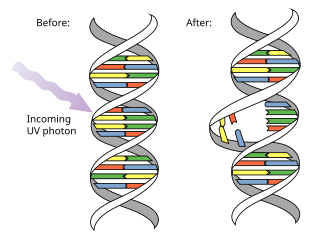
Pyrimidine dimers are molecular lesions formed from thymine or cytosine bases in DNA via photochemical reactions, commonly associated with direct DNA damage. Ultraviolet light induces the formation of covalent linkages between consecutive bases along the nucleotide chain in the vicinity of their carbon–carbon double bonds. The photo-coupled dimers are fluorescent. The dimerization reaction can also occur among pyrimidine bases in dsRNA —uracil or cytosine. Two common UV products are cyclobutane pyrimidine dimers (CPDs) and 6–4 photoproducts. These premutagenic lesions alter the structure of the DNA helix and cause non-canonical base pairing. Specifically, adjacent thymines or cytosines in DNA will form a cyclobutane ring when joined together and cause a distortion in the DNA. This distortion prevents replication or transcription machinery beyond the site of the dimerization. Up to 50–100 such reactions per second might occur in a skin cell during exposure to sunlight, but are usually corrected within seconds by photolyase reactivation or nucleotide excision repair. In humans, the most common form of DNA repair is nucleotide excision repair (NER). In contrast, organisms such as bacteria can counterintuitively harvest energy from the sun to fix DNA damage from pyrimidine dimers via photolyase activity. If these lesions are not fixed, polymerase machinery may misread or add in the incorrect nucleotide to the strand. If the damage to the DNA is overwhelming, mutations can arise within the genome of an organism and may lead to the production of cancer cells. Uncorrected lesions can inhibit polymerases, cause misreading during transcription or replication, or lead to arrest of replication. It causes sunburn and it triggers the production of melanin. Pyrimidine dimers are the primary cause of melanomas in humans.
Missense mRNA is a messenger RNA bearing one or more mutated codons that yield polypeptides with an amino acid sequence different from the wild-type or naturally occurring polypeptide. Missense mRNA molecules are created when template DNA strands or the mRNA strands themselves undergo a missense mutation in which a protein coding sequence is mutated and an altered amino acid sequence is coded for.

Apolipoprotein B mRNA editing enzyme, catalytic polypeptide 1 also known as C->U-editing enzyme APOBEC-1 is a protein that in humans is encoded by the APOBEC1 gene.

Cytidine deaminase is an enzyme that in humans is encoded by the CDA gene.

DNA dC->dU-editing enzyme APOBEC-3F is a protein that in humans is encoded by the APOBEC3F gene.

DNA repair protein REV1 is a protein that in humans is encoded by the REV1 gene.

Protein reversionless 3-like (REV3L) also known as DNA polymerase zeta catalytic subunit (POLZ) is an enzyme that in humans is encoded by the REV3L gene.

Apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3A, also known as APOBEC3A, or A3A is a gene of the APOBEC3 family found in humans, non-human primates, and some other mammals. It is a single-domain DNA cytidine deaminase with antiviral effects. While other members of the family such as APOBEC3G are believed to act by editing ssDNA by removing an amino group from cytosine in DNA, introducing a cytosine to uracil change which can ultimately lead to a cytosine to thymine mutation, one study suggests that APOBEC3A can inhibit parvoviruses by another mechanism. The cellular function of APOBEC3A is likely to be the destruction of foreign DNA through extensive deamination of cytosine.Stenglein MD, Burns MB, Li M, Lengyel J, Harris RS. "APOBEC3 proteins mediate the clearance of foreign DNA from human cells". Nature Structural & Molecular Biology. 17 (2): 222–9. doi:10.1038/nsmb.1744. PMC 2921484. PMID 20062055.

Probable DNA dC->dU-editing enzyme APOBEC-3B is a protein that in humans is encoded by the APOBEC3B gene.

APOBEC is a family of evolutionarily conserved cytidine deaminases.

DNA dC->dU-editing enzyme APOBEC-3H, also known as Apolipoprotein B mRNA-editing enzyme catalytic polypeptide-like 3H or APOBEC-related protein 10, is a protein that in humans is encoded by the APOBEC3H gene.
Somatic hypermutation is a cellular mechanism by which the immune system adapts to the new foreign elements that confront it, as seen during class switching. A major component of the process of affinity maturation, SHM diversifies B cell receptors used to recognize foreign elements (antigens) and allows the immune system to adapt its response to new threats during the lifetime of an organism. Somatic hypermutation involves a programmed process of mutation affecting the variable regions of immunoglobulin genes. Unlike germline mutation, SHM affects only an organism's individual immune cells, and the mutations are not transmitted to the organism's offspring. Because this mechanism is merely selective and not precisely targeted, somatic hypermutation has been strongly implicated in the development of B-cell lymphomas and many other cancers.
DNA Polymerase V is a polymerase enzyme involved in DNA repair mechanisms in bacteria, such as Escherichia coli. It is composed of a UmuD' homodimer and a UmuC monomer, forming the UmuD'2C protein complex. It is part of the Y-family of DNA Polymerases, which are capable of performing DNA translesion synthesis (TLS). Translesion polymerases bypass DNA damage lesions during DNA replication - if a lesion is not repaired or bypassed the replication fork can stall and lead to cell death. However, Y polymerases have low sequence fidelity during replication. When the UmuC and UmuD' proteins were initially discovered in E. coli, they were thought to be agents that inhibit faithful DNA replication and caused DNA synthesis to have high mutation rates after exposure to UV-light. The polymerase function of Pol V was not discovered until the late 1990s when UmuC was successfully extracted, consequent experiments unequivocally proved UmuD'2C is a polymerase. This finding lead to the detection of many Pol V orthologs and the discovery of the Y-family of polymerases.
Mutational signatures are characteristic combinations of mutation types arising from specific mutagenesis processes such as DNA replication infidelity, exogenous and endogenous genotoxin exposures, defective DNA repair pathways, and DNA enzymatic editing.














